Proceedings of the 36th Meeting
Working Group on Prolamin Analysis
and Toxicity (PWG)
Carmen Gianfrani and Peter Koehler
January 2024
Preface
After hybrid and online meetings from 2020 to 2022 due to the global Covid 19 pandemic, it was again possible to organise a physical meeting of the Working Group on Prolamin Analysis and Toxicity (PWG) in 2023. The 36th meeting of the PWG was held from 21st to 23rd September 2023 in Wageningen, The Netherlands. Initially, the meeting was already planned for autumn 2021. The meeting was held at Wageningen International Congress Centre B.V (WICC). The local hosts René Smulders, Ingrid van der Meer, and Peter Weegels were present during the entire meeting. René and Ingrid were also available after the official programme and organised a joint dinner with the participants that stayed for another day after the meeting. Daniëlle van der Wee from plant breeding at Wageningen University & Research did a fantastic job in running the conference office. Thank you very much! 39 Persons participated in the meeting. Apart from the group members, the audience comprised four invited speakers, guests from academia, industry, and international coeliac societies. Representatives from cereal starch producers, producers of gluten-free foods, as well as manufacturers of kits for gluten analysis participated from industry. Analytical and clinical work in the field of coeliac disease, gluten intolerances, gluten and wheat breeding were presented in ten talks. The symposium <Dissecting the pathogenesis of coeliac disease using organoids= comprised four presentations of invited speakers. The meeting was concluded by a talk on regulatory aspects of gluten analysis and labelling and a statement from a starch producer on gluten legislation. After the sessions, there was enough time for discussions. I would like to express my thanks to all participants of the meeting for their active contributions and the open discussions. I am in particular grateful to René Smulders and his team for their enthusiasm and hospitality. Finally, I would like to say thank you to all friends, colleagues, sponsors and participants for their inspiration and ongoing support of the PWG and the meeting. After 13 years this meeting was the last one for me as the chairman and as a member of the PWG. Carmen Gianfrani follows me in this function, and I am wishing her all the best as the new leader of the group! I would like to express my thanks to all friends, colleagues, speakers, and hosts. You contributed to the scientific progress in the field of gluten and coeliac disease. Last but not least, I am grateful for wonderful meetings and meeting locations during the last years. I enjoyed working with you and hope that the PWG will remain a unique group of scientists from different disciplines dealing with all aspects of gluten related intolerances and hypersensitivities.
Esslingen, January 2024
Peter Koehler
Executive Summary
Sixteen presentations covered all aspects related to gluten, coeliac disease (CD) and other relevant hypersensitivities, as well as amylase-trypsin inhibitors (AT) and legal issues. All authors have sent abstracts that are compiled in this proceedings book.
Analytical session
This session comprised four talks. In the first presentation, the benefits and limitations of wheat proteomics were discussed. Acquiring quantitative information from peptide level into protein level is most important but still a challenge. Another presentation on proteomics showed that low allergen and low immunogenicity wheats can be bred and produced in larger quantities and economically acceptable costs. A study on the knock-out of amylase-trypsin inhibitors (ATI) showed that it was possible to generate ATI-depleted wheat, but low ATI lines had not necessarily low bioactivity. Finally, coeliac disease epitopes were eliminated from wheat gluten by the CRISPR/Cas technology. This could yield wheat varieties that would fit into the niche of low-gluten products for people with non-coeliac gluten (or wheat) sensitivity.
Clinical session
This session included six presentations. In particular, interleukins (IL) -10 and -2 play an important role in CD research. IL 10 is involved in the transition from potential to overt CD. IL-2 is only found in CD patients and is a powerful biomarker after gluten ingestion and in the development of CD. These considerations were supported by another study showing that metabolomics and the determination of CD-associated antibodies and cytokines can help in predicting CD. Transglutaminase 2 (TG2) inhibition is a promising therapy of CD. A candidate molecule is currently tested in a phase 2b study. Finally, the last presentation in this session highlighted the role of pyroptosis and necroptosis in the release of pro-inflammatory components expanding inflammation and tissue damage.
Symposium: Dissecting the pathogenesis of coeliac disease using organoids
The symposium included four presentations of invited speakers. An induced pluripotent stem cell (iPSC)-derived Intestine-Chip was developed to mimic the cell populations of the intestinal mucosa that can be used to study CD pathogenesis in vitro under in vivo conditions. An autologous co-culture system is generated to monitor key aspects of the CD mucosal immune response. The last two presentations were on intestinal organoids that closely mimic the in vivo situation and can be used to study the pathomechanism of CD. They are isolated from endoscopic or surgical mucosa-samples and carry the epithelial properties of the human individual they were collected from, including the disease determining alterations.
Analytical research reports
Proteomics analysis of wheat gluten: how do we interpret and summarise the data?
Twan America
BU Bioscience, Wageningen Plant Research, WUR, Wageningen, the Netherlands
Abstract
Proteomics characterisation of complex protein extracts is mostly performed using the so-called <bottom-up= approach. In this approach, the extract of proteins is digested with a specific protease, in most cases being trypsin but in the case of gluten, chymotrypsin is preferred. This results in a highly complex mixture of peptides. This mixture is then separated on an LC-MS system, where the peptides are first separated by reversed phase chromatography and directly from the elution of the column sprayed into a mass spectrometer. There are different options for MS detection, identification and quantitation of peptides. Non-targeted data-dependent (DDA) or data-independent analysis (DIA) provide the most holistic approach of characterizing the complexity of the sample. In both cases there are several steps in data processing in which MS signal information is deconvoluted, integrated and combined into (semi-)quantitative information at either peak, peptide, or protein level.
I presented some examples where I indicated the advantages and limitations of the different steps of data integration. In summary:
• The complete peak pattern information gives most accurate and complete quantitative info of all detected components. It can be used for classification and difference analysis but does not (yet) provide identity information.
• The information gathered at peptide sequence level is very detailed, often based on agglomerating multiple peaks signals and as such also highly quantitative. However as not all peaks are identified, there are missing data in the peptide tables, especially at low intensity signals. Depending on the parameters of the database search algorithm some modifications of peptides may be recognised or missed. Unexpected modifications of peptides are mostly overlooked.
• The information integrated at individual protein level is based on the sequence information that was provided as the protein sequence database input for the identification algorithm. Combining multiple peptide quantitative info to the protein level is highly dependent on the correct coverage of protein sequences in the database. If a high level of redundancy or a large number of protein isoforms are present (as is the case for most gluten proteins) this will result in a complex puzzle for resolving which peptides belong to which protein isoform. This process is error prone and highly dependent on the used algorithm and input data.
• Integrating quantitative information from peptide level into protein class level is probably the most integrated, accurate and relevant type of information, but requires manual classification of proteins, as this class information itself is not (well) provided in the Uniprot database.
Quantitative proteomics of more than 2000 proteins from different wheat species and potential allergenicity
Detlef Schuppan1,2, Muhammad Afzal3, Malte Sielaff4, Manjusha Neerukonda1, Ute Distler4, Stefan Tenzer4, Friedrich H. Longin3
1 Institute of Translational Immunology and Research Center for Immune Therapy (FZI), University Medical Center of the Johannes Gutenberg University Mainz, Mainz, Germany 2 Division of Gastroenterology, Beth Israel Deaconess Medical Center, Harvard Medical School, Boston, MA, USA 3 State Plant Breeding Institute, University of Hohenheim, Stuttgart, Germany 4 Institute for Immunology and Research Center for Immune Therapy (FZI), University Medical Center of the Johannes Gutenberg University Mainz, Mainz, Germany All authors contributed equally.
Abstract
We define non coeliac wheat sensitivity (NCWS) as an inflammatory reaction to wheat with intestinal, extra-intestinal and also central nervous system reactions. Wheat amylase trypsin inhibitors (ATI), which represent 3-4% of wheat proteins are activators of intestinal myeloid cells via toll like receptor 4, which exacerbate chronic inflammatory, and often autoimmune diseases in mouse models [1-9] and clinical studies [10-13]. Moreover, abdominal complaints and skin problems are often triggered by allergic reactions to (mainly non-gluten) wheat proteins. Here, a clinically delayed type 2 allergy to wheat is a major cause of irritable bowel syndrome (a condition that affects 10-15% of most populations) [14-15]. We used liquid chromatography-tandem mass spectrometry followed by high-end label-free proteomics on quantitative protein extracts from five wheat species (diploid einkorn, tetraploid emmer, and durum, hexaploid spelt and modern wheat, each with 10 cultivars grown in three diverse environments (150 samples). At least 2540 proteins could be identified in each species. More than 50% of proteins differed between species and many of these proteins are implicated in dough quality, plant stress regulation, grain-starch synthesis, and 3 importantly 3 classical IgE-mediated allergies. Genetics and environmental factors were major determinants of the species-specific protein patterns observed. Notably, einkorn expressed 5.4-fold lower quantities of potential allergens and 7.2-fold lower quantities of classical immune-stimulatory ATI than the hexaploid wheats, and the tetraploid wheats showed intermediate values. Our data show that low allergen and low immunogenicity wheats can be bred and grown in a targeted proteome-based approach, and also produced in larger quantities and economically acceptable costs [16].
References
1. Junker Y, et al. Wheat amylase trypsin inhibitors drive intestinal inflammation via activation of toll-like receptor 4. J Exp Med 2012; 209: 2395-2408.
2. Zevallos VF, et al. Nutritional Wheat Amylase-Trypsin Inhibitors Promote Intestinal Inflammation via Activation of Myeloid Cells. Gastroenterology 2017; 152: 110031113.e12.
3. Ashfaq-Khan M, et al. Dietary wheat amylase trypsin inhibitors promote features of murine non-alcoholic fatty liver disease. Sci Rep 2019; 9: 1-14.
4. Bellinghausen I, et al. Wheat amylase-trypsin inhibitors exacerbate intestinal and airway allergic immune responses in humanized mice. J Allergy Clin Immunol 2019; 143: 201-212.e4.
5. Zevallos VF, et al. Dietary wheat amylase trypsin inhibitors exacerbate murine allergic airway inflammation. Eur J Nutr 2019; 58: 1507-1514.
6. Caminero A, et al. Lactobacilli Degrade Wheat Amylase Trypsin Inhibitors to Reduce Intestinal Dysfunction Induced by Immunogenic Wheat Proteins. Gastroenterology 2019; 156: 2266-2280.
7. Pickert G, et al. Wheat Consumption Aggravates Colitis in Mice via Amylase Trypsin Inhibitor-mediated Dysbiosis. Gastroenterology 2020; 159: 257-272.e17.
8. Dos Santos Guilherme M, et al. Dietary Wheat Amylase Trypsin Inhibitors Impact Alzheimer's Disease Pathology in 5xFAD Model Mice. Int J Mol Sci 2020; 31: 6288.
9. Zevallos VF, et al. Dietary wheat amylase trypsin inhibitors exacerbate CNS inflammation in experimental multiple sclerosis. Gut 2023; 73: 5-6.
10. Carroccio, A, et al. Wheat Consumption Leads to Immune Activation and Symptom Worsening in Patients with Familial Mediterranean Fever: A Pilot Randomized Trial. Nutrients 2020; 12: 1127.
11. Engel S, et al. Attenuation of immune activation in patients with multiple sclerosis on a wheat-reduced diet: a pilot crossover trial. Ther Adv Neurol Disord 2023; 16: 17562864231170928. Epub ahead of print.
12. Liwinski T, et al. A prospective pilot study of a gluten-free diet for primary sclerosing cholangitis and associated colitis. Aliment Pharmacol Ther 2023; 57: 224-236.
13. Armandi A, et al. Short term gluten-free diet in patients with Non-Alcoholic Fatty Liver Disease: a proof of concept randomized, placebo-controlled study. Nutrients, in press.
14. Fritscher-Ravens A, et al. Confocal Endomicroscopy Shows Food-Associated Changes in the Intestinal Mucosa of Patients With Irritable Bowel Syndrome. Gastroenterology 2014; 147: 1012-1020.
15. Fritscher-Ravens A, et al. Many Patients With Irritable Bowel Syndrome Have Atypical Food Allergies Not Associated With Immunoglobulin E. Gastroenterology 2019; 157: 109-118.
16. Afzal M, et al. Reference proteomes of five wheat species as starting point for future design of cultivars with lower allergenic potential. NPJ Sci Food 2023;7: 9.
Knock-out of ATI in wheat and resulting bioactivity
Manjusha Neerukonda1, Marco Bonarrigo2, Arianna Frittelli2, Francesco Sestili2, Francesca Fayer3, Pasquale Mansueto3, Antonio Carroccio3, Stefania Masci2, Detlef Schuppan1,4
1 Institute of Translational Immunology, University Medical Center, Mainz, Germany 2 Department of Agriculture and Forestry Science, University of Tuscia, Viterbo, Italy 3 Internal Medicine Unit, <V Cervello Hospital=, Department of Health Promotion Sciences, Maternal and Infant Care, Internal Medicine and Medical Specialties (PROMISE), University of Palermo, Italy 4 Division of Gastroenterology, Beth Israel Deaconess Medical Center, Harvard Medical School, Boston, MA, USA
Abstract
Wheat, the major staple food, with its several proteins that are known to be the triggers of allergic reactions, cause intestinal and extra- intestinal inflammatory diseases, such as coeliac disease and non-coeliac wheat sensitivity. Alpha-amylase/trypsin inhibitors (ATIs), a family of several non-gluten wheat proteins, encoded by a multigene family dispersed over different chromosomes, are the central allergens in baker9s asthma, especially tetrameric ATIs CM3 and CM16, and dimeric ATI 0.28. ATIs also engage the CD14-MD2-TLR4 (toll-like receptor 4) complex on myeloid cells and trigger an innate immune response that promotes intestinal and extra-intestinal diseases.
With the aim of obtaining wheat plants with decreased expression of the specific ATI proteins CM3, CM16, and 0.28, we produced bread wheat lines with CM3, CM16 and 0.28 ATI genes silenced by means of RNAi (RNAi lines), and durum wheat lines with CM3 and CM16 ATI genes silenced by genome editing using CRISPR/Cas9 (GE lines).
Quantitative ATI extracts from flours, depleted of LPS using polymyxin B affinity chromatography, were tested for their in vitro TLR4-stimulating bioactivity using a Hela TLR4 dual luciferase reporter cell line that produces dose-dependent luminescence signals. Two RNAi silenced lines resulted in increased bioactivity, while the two GE lines showed a significant reduction of bioactivity compared to their respective wild-type reference genotypes. RNAi lines exhibited several pleiotropic effects, varying from non-specific silencing of high molecular weight glutenin subunits to overexpression of ÷- and ÷-gliadins and other proteins with trypsin inhibition activity. No marked changes in seed composition were observed in the GE lines.
Overall, GE proved to be a powerful genome modification tool. However, in vitro bioactivity readouts suggest a need to compare different knockout of ATI sub-species in different wheat varieties to ascertain that NCWS patients may benefit from ATI-depleted wheat.
Impact analysis of low-gluten, CD-safe wheat
Marinus J. M. (René) Smulders
Plant Breeding, Wageningen University & Research, Wageningen, The Netherlands
Abstract
In wheat, coeliac disease (CD) epitopes occur mostly in gliadins, while the baking quality is determined predominantly by glutenins. Thus, mutating CD epitopes from gliadin genes, and removing some gliadin genes altogether, can be used to lower the immunogenicity of wheat. Unfortunately, bread wheat varieties contain around 100 gliadin and glutenin genes, most of which contain one or more CD epitopes. The genes are genetically linked on chromosomes 1 and 6 of wheat. For that reason, plant breeding cannot generate bread wheat that is safe for coeliac disease patients while retaining baking quality solely by combining natural or randomly induced mutations in gliadins and glutenins. However, in combination with targeted mutagenesis by gene editing with CRISPR/Cas it is feasible [1,2,3]. Recent improvements in the regeneration of wheat [4] make it realistic to apply in commercial germplasm. The major epitopes are the primary targets. Selection and screening at DNA and protein level must be confirmed with T cell tests, etc. [5]. Once hypoallergenic loci of multiple genes have been obtained, they may be combined through regular crossing and selecting. An intermediate product will be low-gluten wheat varieties. These varieties are noy yet safe for CD patients, but they may fit into the niche of low-gluten products for people who want to lower their gluten intake, e.g., people with (self-diagnosed) non-coeliac gluten (or wheat) sensitivity.
In the EU, plants for which, during the breeding process, gene editing has been used for targeted mutagenesis, are regulated as GMOs [6]. The European Commission, asked by the Council of Member States, concluded in 2021 that the Directive 2001/18/EC on the deliberate release of GMOs into the environment, was <not fit for purpose= anymore. The Commission published their proposal for a revised regulation in July 2023 [7]. As part of the process of developing a new regulation, an inception impact assessment was made of the socio-economic impacts, including the potential contribution of low-gluten, CD-safe wheat for food security, nutrition, and public health [8].
References
1.Jouanin A, Gilissen LJWJ, Boyd LA, et al. (2018) Food processing and breeding strategies for coeliac-safe and healthy wheat products. Food Res Int 110: 11321. https://doi.org/10.1016/j.foodres.2017.04.025
2. Sánchez-León S, Gil-Humanes J, Ozuna CV, Giménez MJ, Sousa C, Voytas DF, Barro F (2018) Low-gluten, nontransgenic wheat engineered with CRISPR/Cas9. Plant Biotechnol J 16: 902-910. https://doi.org/10.1111/pbi.12837
3. Jouanin A, Schaart JG, Boyd LA, Cockram J, Leigh FJ, Bates R, Wallington EJ, Visser RGF, Smulders MJM. Outlook for coeliac disease patients: Towards bread wheat with hypoimmunogenic gluten by gene editing of ³- and ³-gliadin gene families. BMC Plant Biol 2019; 19: 333. https://doi.org/10.1186/s12870-019-1889-5 .
4. Wang K, Shi L, Liang X, et al. The gene TaWOX5 overcomes genotype dependency in wheat genetic transformation. Nat Plants 2022; 8: 110-117. https://doi.org/10.1038/s41477-021-01085-8
5. Jouanin A, LJWJ Gilissen, JG Schaart, et.al. CRISPR/Cas9 gene editing of gluten in wheat to reduce gluten content and exposure 3 reviewing methods to screen for coeliac safety. Front Nutrit 2020; 7: 51. https://doi.org/10.3389/fnut.2020.00051 .
6. Jouanin A, Boyd LA, Visser RGF, Smulders MJM. Development of wheat with hypoimmunogenic gluten obstructed by the gene editing policy in Europe. Front Plant Sci 2018; 9: 1523. https://doi.org/10.3389/fpls.2018.01523
7. European Commission. (2023) New Techniques in Biotechnology. https://food.ec.europa.eu/plants/genetically-modified-organisms/new-techniques-biotechnology_en#ongoing-mandates
8. Sánchez B, Barro F, Smulders MJM, Gilissen LJWJ, Rodríguez Cerezo E. Socioeconomic impact of low-gluten, celiac-safe wheat developed through gene editing. 2020; EUR 31380 EN, Publications Office of the European Union, Luxembourg. https://doi.org/10.2760/280847
5. Clinical research reports
Regulatory mechanisms controlling in the gut mucosa the transition from potential to overt coeliac disease
Serena Vitale1, Ilaria Mottola1, Renata Auricchio2, Silvia Gregori3, Riccardo Troncone2, Carmen Gianfrani1
1 Institute of Biochemistry and Cell Biology, CNR, Naples, Italy 2 Department of Medical Translational Sciences and European Laboratory for the Investigation of Food-Induced Diseases, University Federico II, Naples, Italy 3 Mechanisms of Peripheral Tolerance Unit, San Raffaele Telethon Institute for Gene Therapy (SR-Tiget), IRCCS San Raffaele Scientific Institute, Milan, Italy
Abstract
The main feature of Coeliac Disease (CD) pathogenesis is an altered immune response upon ingestion of gluten containing food. Gluten-reactive T cells infiltrating the small intestinal mucosa have a key pathogenic role, due to a massive production of interferon(IFN)-÷. To date, several pharmaceutical strategies have been proposed to recover the immune tolerance to gluten, aimed to place, or to support, the gluten-free diet therapy.
Regulatory T cells secreting interleukin(IL)-10 (Tr1) are pivotal in controlling the adverse inflammatory reactions induced by dietary antigens and microbes in intestinal tract, thus contributing to the intestinal homeostasis [1]. We reported that IL-10 is markedly produced in coeliac intestinal mucosa, although the ratio of IL-10/IFN-÷ amount in the gut mucosa of acute CD patients is significantly lower compared to the level found in healthy controls and treated CD patients [2]. The treatment with exogenous IL-10 of coeliac intestinal mucosa in organ culture models prevents the immune activation induced by gluten challenge. We have also demonstrated that CD intestinal mucosa harbours Tr1 cells that significantly inhibit the inflammatory T cell response to gluten [3].
We also demonstrated that IL-10 modulates the gluten-induced inflammatory reaction in the gut mucosa of children with potential CD, a condition characterised by positive anti-tissue transglutaminase/anti-endomysium serology but absence of villous atrophy [4]. The discovery of CD49b and LAG-3 as specific cell surface markers of Tr1 cells allows direct monitoring and quantification of this cell subsets in peripheral blood and in gut mucosa [5]. Notably, gluten-reactive Tr1 cells can be differentiated in vitro upon co-culture of naïve T cells with tolerogenic dendritic cells (DC-10), a peculiar subset of DC characterised by high production of IL-10. Interestingly we found an increased frequency of IL-10-secreting DC (DC-10) and of IL-10-secreting Tr1 cells specific for gliadin in potential-CD patients [6].
Collectively, these findings strongly suggest that IL-10 and Tr1 cells have a role in controlling the natural transition from potential- to acute-CD, and open suitable perspective for a cell-based therapy to restore the immune tolerance to gluten in CD.
References
1. Roncarolo MG, et al. The Biology of T Regulatory Type 1 Cells and Their Therapeutic Application in Immune-Mediated Diseases. Immunity 2018; 9(6): 1004-1019.
2. Salvati V.M, et al. Recombinant human interleukin 10 suppresses gliadin dependent T cell activation in ex vivo cultured coeliac intestinal mucosa. Gut 2005; 54: 46-53.
3. Gianfrani C, et al. Gliadin-specific type 1 regulatory T cells from the intestinal mucosa of treated celiac patients inhibit pathogenic T cells. J Immunol 2006; 177: 4178-4186.
4. Camarca A, et al. Gliadin-reactive T cells in Italian children from preventCD cohort at high risk of celiac disease. Pediatr Allergy Immunol 2017; 28: 362-369.
5. Gagliani N, et al. Coexpression of CD49b and LAG-3 identifies human and mouse T regulatory type 1 cells. Nature Medicine 2013; 19: 739-746.
6. Passeri L, et al. Tolerogenic IL-10-engineered dendritic cell-based therapy to restore antigen-specific tolerance in T cell mediated diseases. J Autoimmun 2023; 138: 103051.
Paediatric screening for type 1 diabetes and coeliac disease in Italy
Carlo Catassi
Department of Paediatrics, Polytechnic University of Marche, Ancona, Italy
Abstract
On September 17th, 2023, the Italian Parliament unanimously approved Law n. 130/2023 introducing the nationwide screening for type 1 diabetes (T1D) and coeliac disease (CD) in the general population aged 1-17 years, as part of a new public health programme aimed to reduce the impact of these chronic diseases [1].
The specific aims of the new Italian Law are: 1) the identification of children and adolescents during their pre-symptomatic phase of T1D. This stage is an opportunity for education, awareness and other programmes that can help to prevent the potentially lethal diabetic ketoacidosis associated with late clinical diagnosis, and to receive disease modifying therapies that can prevent the onset of clinical disease or delay its progression; 2) the early diagnosis of atypical or silent CD currently escaping clinical diagnosis (the invisible part of the so-called <coeliac iceberg=), in order to allow prompt treatment with the GFD and prevention of long-term complications, such as short stature, osteoporosis, infertility, gut lymphoma, and small intestine adenocarcinoma [2,3].
General screening for both T1D and CD is possible thanks to the availability of specific and sensitive serological biomarkers that can be measured on a few drops of whole blood taken by a lancing device. During the last few decades, a major achievement in T1D research has been the identification of a long pre-symptomatic phase that is marked by the appearance of disease-specific serum autoantibodies. These are collectively defined as islet autoantibodies and include four specificities (against glutamic acid decarboxylase - GAD -, insulin, IA-2 and ZnT8). In children and adolescents, the positivity for g2 islet autoantibodies is associated with the greatest risk (almost certainty) of future disease; positivity for one autoantibody indicates an intermediate risk, while no risk is associated with a negative autoantibody status. Likewise, the development of CD, either typical or clinically atypical/silent, is heralded by the finding of specific serological markers, i.e. serum IgA class tissue transglutaminase (TTG) and endomysial autoantibodies (EMA). T1D and CD-specific autoantibodies can be tested together in combined screening procedures, as indicated by the new Italian Law.
The implementation of Law n.130/2023 raises several challenging questions. For instance, it will be necessary to make a choice about the age of screened children, to select the appropriate autoantibody measurement assays and complementary genetic tests, to choose between venous vs capillary blood sampling or use of dried blood spots. The psychological impact of screening in children and families needs careful evaluation as well. Finally, education programmes and follow-up protocols will be required for individuals identified at-risk, and interventions for disease modification and prevention need to be defined (risk stratification, outcome measures, timing, and duration). Despite these challenges, a new and exciting scenario in public health has been uncovered, with Law 130/2023 representing a milestone in the history of paediatric preventive medicine in Italy and a possible reference model for other countries.
References
1. Available at https://www.gazzettaufficiale.it/eli/id/2023/09/27/23G00140/sg
2. Quattrin T, Mastrandrea LD, Walker LSK. Type 1 diabetes. Lancet 2023; 401(10394): 2149-2162. doi: 10.1016/S0140-6736(23)00223-4
3. Catassi C, Verdu EF, Bai JC, Lionetti E. Coeliac disease. Lancet 2022; 399(10344): 2413-2426. doi: 10.1016/S0140-6736(22)00794-2
Can coeliac disease be predicted?
Riccardo Troncone, Renata Auricchio, Luigi Greco
Department of Medical Translational Sciences and European Laboratory for the Investigation of Food-Induced Diseases, University Federico II, Naples, Italy
Abstract
Prospective studies of cohorts of infants at risk of developing coeliac disease have recently been developed [1]. They are based on longitudinal collection of clinical data and biological samples. The identification of risk factors and biological markers predicting the onset of the disease are the main outcome of such studies. Both genetic and environmental risk factors have been identified. Family history, HLA haplotypes, female sex are all important contributors [2]. Children homozygous for HLA DQ2 are significantly more prone to develop CD and the risk is much increased in females [2]. Environmental factors include early life infections, particularly by enterovirus. Among dietary factors high early life cumulative gluten intake has emerged as risk factor, while early suggestions of a protective effect of breast feeding and a possible role of timing of gluten introduction have not been confirmed [2]. Mediterranean-like diet has been associated with a low risk of CD autoimmunity: consumption of greater amounts of carbohydrates, particularly starch and sugars, and lower amounts of legumes, vegetables, fruits, and milk products were reported in children who developed CD [3].
The longitudinal collection of samples followed by the comparison of infants developing CD with those remaining healthy during follow-up has allowed the identification of predictive biomarkers. Among genetic factors the expression of candidate genes was analysed at various timepoints and hyperexpression found months before diagnosis [4]. Circulating miRNA were also detected in circulation before seroconversion [5]. Metabolomics studies also contributed: children progressing to CD showed increased amounts of triacylglycerol and a decreased levels of phosphatidylcholine by age of three months as compared to controls. Finally, CD-associated antibodies and cytokines were investigated: early and isolated production of antigliadin IgA antibody was noted in those who did not progress, while changes in cytokine pattern were found to precede the appearance of anti-tissue transglutaminase antibodies [6,7].
CD is a multifactorial disease. Prospective studies of at-risk infants have shed light on the natural history of this condition. Biomarkers sign the different stages of the disease helping to predict those at risk of progression and amenable to prevention strategies.
References
1. Auricchio R, Troncone R. Can Celiac Disease Be Prevented? Front Immunol 2021; 12: 672148.
2. Meijer CR, et al. Prediction Models for Celiac Disease Development in Children From High-Risk Families: Data From the PreventCD Cohort. Gastroenterology 2022; 163: 426-436.
3. Auricchio R, et al. Gluten consumption and inflammation affect the development of celiac disease in at-risk children. Sci Rep 2022; 12: 5396.
4. Galatola M, et al. Presymptomatic Diagnosis of Celiac Disease in Predisposed Children: The Role of Gene Expression Profile. J Pediatr Gastroenterol Nutr 2017; 65: 314-320.
5. Tan IL, et al. Circulating miRNAs as Potential Biomarkers for Celiac Disease Development. Front Immunol 2021; 12: 734763.
6. Auricchio R, et al. A Phospholipid Profile at 4 Months Predicts the Onset of Celiac Disease in at-Risk Infants. Sci Rep 2019; 9: 14303.
7. Auricchio R, et al. Antibody Profile, Gene Expression and Serum Cytokines in At-Risk Infants before the Onset of Celiac Disease. Int J Mol Sci 2023; 24: 6836.
Interleukin-2 readouts in coeliac disease
Knut E. A. Lundin1,2
Norwegian Coeliac Disease Research Centre, Institute of Clinical Medicine, Faculty of Medicine, University of Oslo, Oslo, Norway 2 Department of Gastroenterology, Oslo University Hospital Rikshospitalet, Oslo, Norway
Abstract
Interleukin-2 (IL-2) is a secreted cytokine protein with molecular weight 15,5-16 kDa and produced by CD4+ and CD8+ T cells exclusively [1]. It stimulates the growth of helper, cytotoxic and regulatory T cells. It belongs to a family of similar cytokines encompassing IL-4, IL-7, IL-19, IL-15 and IL-21. The IL-2 receptors consist of three different chains; the alpha receptor CD25 is of low affinity, the beta receptor CD122 and the gamma receptor CD132. It is involved in differentiation of memory T cells and regulatory T cells and increases action of natural killer cells and cytotoxic T cells. Clinical use was investigated in cancer treatment, but its use was limited due to flu-like symptoms, nausea/vomiting and diarrhoea, weakness and drowsiness.
A major break-through in the understanding of coeliac disease (CD) came when Anderson and colleagues showed that exposure of gliadin peptides led to release of a range of cytokines with especially IL-2, but also IL-8 and IL-10 [2]. These cytokines were found both after oral and parenteral (intradermal) exposure. The release of IL-2 is important, as many investigators have focused on the innate immune, and not the adaptive, system as the most important part of the rapid response to gluten. IL-2 was found after 2 hours, peaked at 4-6 hours, and correlated with symptoms like nausea and vomiting. Large inter-individual differences between CD patients are observed [3, 4]. The most complete picture was described by Tye-Din who challenged 295 adult CD patients, where he found that HLA-DQ2 homozygosity and previous challenge gave highest IL-2 levels [5]. Secretion of IL-2 is only found in CD and not in <non-coeliac gluten sensitivity= [6]. The amount of IL-2 largely correlates with activation of gliadin-specific T cell activation measured by HLA-DQ2:gluten tetramers [7, 8]. The amount of gluten ingested correlates with IL-2 response [9]. IL-2 response holds promise to be used in clinical trials of drugs for CD [10]. Measurement of IL-2 in serum is dependent on supersensitive assays. The Mesoscale platform is mostly used. The assay accuracy and inter-individual variety of response remains challenging in diagnostic and therapeutic settings.
References
1. Abbas AK, et al. Revisiting IL-2: Biology and therapeutic prospects. Sci Immunol 2018; 3(25): eaat1482.
2. Goel G, et al. Cytokine release and gastrointestinal symptoms after gluten challenge in celiac disease. Sci Adv, 2019; 5(8): eaaw7756.
3. Tye-Din JA, et al. Elevated serum interleukin-2 after gluten correlates with symptoms and is a potential diagnostic biomarker for coeliac disease. Aliment Pharmacol Ther 2019; 50(8): 901-910.
4. Goel G, et al. Serum cytokines elevated during gluten-mediated cytokine release in coeliac disease. Clin Exp Immunol 2020; 199(1): 68-78.
5. Tye-Din JA, et al. Patient factors influencing acute gluten reactions and cytokine release in treated coeliac disease. BMC Med 2020; 18(1): 362.
6. Tye-Din JA, et al. Cytokine release after gluten ingestion differentiates coeliac disease from self-reported gluten sensitivity. United European Gastroenterol J, 2020; 8(1): 108-118.
7. Sarna VK, et al. HLA-DQ:gluten tetramer test in blood gives better detection of coeliac patients than biopsy after 14-day gluten challenge. Gut 2018; 67(9): 1606-1613.
8. Zuhlke S, et al. CD38 expression on gluten-specific T cells is a robust marker of gluten re-exposure in coeliac disease. United European Gastroenterol J 2019; 7(10): 1337-1344.
9. Leonard MM, et al. Evaluating Responses to Gluten Challenge: A Randomized, Double-Blind, 2-Dose Gluten Challenge Trial. Gastroenterology 2021; 160(3): 720-733.e8.
10. Murray JA, et al. Safety and tolerability of KAN-101, a liver-targeted immune tolerance therapy, in patients with coeliac disease (ACeD): a phase 1 trial. Lancet Gastroenterol Hepatol 2023; 8(8): 735-747.
Transglutaminase inhibition in coeliac disease: progress in understanding and clinical studies
Detlef Schuppan1,2,3
1 Institute of Translational Immunology and Research Center for Immune Therapy (FZI) and 2 Clinical Center for Celiac Disease and Autoimmunity, University Medical Center of the Johannes Gutenberg University Mainz, Mainz, Germany 3 Division of Gastroenterology, Beth Israel Deaconess Medical Center, Harvard Medical School, Boston, MA, USA
Abstract
The cellular enzyme transglutaminase 2 (TG2) is both the coeliac disease (CD) autoantigen and a central driver of the pathogenesis of CD [1-4]. It deamidates glutamines of certain dietary gluten peptides in the intestinal mucosa that escape gastrointestinal digestion. The induced change of charge from a neutral glutamine to an acidic glutamate facilitates their antigenic presentation by HLA-DQ2 or HLA-DQ8, and elicits activation and expansion of gluten reactive, mucosa destructive CD4+ T cell clones [5,6]. Blocking the deamidation of gluten peptides should reduce their binding and presentation to gluten reactive T cells, their activation and expansion, and the CD 4 T cell mediated villous atrophy and mucosal inflammation. On this basis, Khosla et al. had developed several small molecules working as TG inhibitors that prevented gluten peptide deamidation, but had limited TG2 specificity and possible toxic side effects [7]. A highly specific TG2-inhibitor (ZED1227) that lacks toxicity and does not inhibit four other major TGs was developed in preclinical and phase 1 clinical studies. In a study of 160 CD patients in remission who were challenged with 3 g of gluten per day for 6 weeks oral ZED1227 prevented gluten-induced mucosal damage and improved patient related outcomes [8-9]. Interestingly, in patients ZED1227 appears to inhibit the enzyme both on the enterocyte surface and in the lamina propria [10]. Currently, ZED1227 is tested in a phase 2b study of 400 patients with mild clinical as well as histological non-diet responsive CD.
References
1. Dieterich W, et al. Identification of tissue transglutaminase as the autoantigen of celiac disease. Nat Med 1997; 3: 797-801.
2. Schuppan D, et al. Celiac disease: from pathogenesis to novel therapies. Gastroenterology 2009; 137: 1912-1933.
3. Lundin KE, Sollid LM. Advances in coeliac disease. Curr Opin Gastroenterol 2014; 30: 154-162.
4. Abadie V, et al. IL-15, gluten and HLA-DQ8 drive tissue destruction in coeliac disease. Nature 2020; 578(7796): 600-604.
5. Molberg O, et al. Tissue transglutaminase selectively modifies gliadin peptides that are recognized by gut-derived T cells in celiac disease. Nat Med 1998; 4: 713-717
6. van de Wal Y, et al. Selective deamidation by tissue transglutaminase strongly enhances gliadin- specific T cell reactivity. J Immunol 1998; 161: 1585-1588.
7. Zhuang R, Khosla C. Substrates, inhibitors, and probes of mammalian transglutaminase 2. Anal Biochem. 2020; 591: 113560.
8. Schuppan D, et al. CEC-3 Trial Group. A Randomized Trial of a Transglutaminase 2 Inhibitor for Celiac Disease. N Engl J Med 2021; 385: 35-45.
9. Büchold C, et al. Features of ZED1227: The First-In-Class Tissue Transglutaminase Inhibitor Undergoing Clinical Evaluation for the Treatment of Celiac Disease. Cells 2022; 11: 1667.
10 Isola J, et al. The Oral Transglutaminase 2 Inhibitor ZED1227 Accumulates in the Villous Enterocytes in Celiac Disease Patients during Gluten Challenge and Drug Treatment. Int J Mol Sci 2023; 24: 10815.
Coexistence of apoptosis, pyroptosis, and necroptosis pathways in Coeliac Disease
Carolina N. Ruera1, Federico Perez1, María Luz Iribarren1, Luciana Guzman2, Lorena Menendez2, Laura Garbi3, Fernando G. Chirdo1
1 Instituto de Estudios Inmunológicos y Fisiopatológicos (IIFP) (UNLP-CONICET-CIC), Facultad de Ciencias Exactas, Universidad Nacional de La Plata, La Plata, Argentina 2 Servicio de Gastroenterología, Hospital de Niños <Sor María Ludovica=, La Plata, Argentina 3 Servicio de Gastroenterología, Hospital San Martin, La Plata, Argentina
Abstract
Regulated cell death incudes different pathways and diverse outcomes for the tissue.
Particularly, apoptosis, considered as an immunologically silent process, is the cell death pathway commonly used for the massive elimination of cells, such as enterocytes in Coeliac Disease (CD). Unlike apoptosis, pyroptosis and necroptosis results in the release of intracellular components with proinflammatory activity, which may expand the inflammation and tissue damage. However, little is known about the role of proinflammatory cell death pathways in human intestinal disease.
The aim of this work was to evaluate the expression of components of proinflammatory cell death pathways in healthy human small intestine and duodenal samples from CD patients.
Duodenal biopsies were collected from active CD patients and non-coeliac individuals during the routine protocol for CD diagnosis in the Gastroenterology Units of the paediatric and adult public hospitals from La Plata city. CD was diagnosed by histological examination of duodenal biopsies and serological assessment. The individuals in the control group (NC) were subjected to endoscopy for reasons other than CD and all were negative for serological or histological evidence of CD. Samples were used for confocal microscopy studies such as TUNEL reaction or Immunofluorescent microscopy (IFI), Western blot (WB), and RT-PCR analysis.
TUNEL reaction showed an increased number of dead cells in the mucosal lamina propria (LP) of CD patients (p<0,002), with most of these being CD138+ plasma cells. Many dying cells expressed FAS and were in close contact with CD3+ T cells, suggesting a possible role for FAS-mediated cytotoxic activity in removal of LP cells. Further analysis showed increases in the expression of active caspase-8 and caspase-3 in CD patients, confirming the activation of apoptosis (p<0.01). In parallel, the biologically active forms of caspase-1, IL-1³ and GSDMD were increased in CD samples indicating the presence of inflammasome-dependent pyroptosis (p<0.01). Necroptosis was also present, as shown by the increase of RIPK3 and phosphorylate MLKL (p-MLKL) (p<0.05). In addition, p-MLKL expression was found in some CD3+ T cells in LP of CD patients and Paneth cells (³defensin+) in the crypts. RT-qPCR analysis revealed that RNAm levels of ZBP1 were also increased in CD patients (p<0.05).
In summary, a high number of dead cells were found in the duodenum of CD patients, with apoptosis, pyroptosis, and necroptosis occurring in parallel. Therefore, in addition to the immunologically silent apoptosis, proinflammatory cell death is also active in enteropathy. DAMPs released during proinflammatory cell death may play a role in the initial steps of the induction of mucosal damage and promote chronic disease. These pathways may contribute to amplify the inflammatory process and damage mechanisms in the intestinal mucosa in CD.
Symposium: Dissecting the pathogenesis of coeliac disease using organoids
Steering epithelial and mesenchymal cell type composition in an iPSC-derived Intestine-Chip
Renée Moerkens1,2, Joram Mooiweer1,2, Aarón D Ramírez-Sánchez1, Roy Oelen1, Cisca Wijmenga1,2, Robert Barrett3,4, Sebo Withoff1,2, Iris Jonkers1
1 Department of Genetics, University Medical Center Groningen, University of Groningen, Groningen, The Netherlands 2 Netherlands Organ-on-Chip Initiative (NOCI), Leiden, The Netherlands 3 Cedars-Sinai Medical Center, Board of Governors Regenerative Medicine Institute, Los Angeles, CA, USA 4 Cedars-Sinai Medical Center, F. Widjaja Foundation Inflammatory Bowel and Immunobiology Research Institute, Los Angeles, CA, USA
Abstract
ntroduction. Growth factor gradients along the crypt-villus axis define the spatial organisation and diversity of intestinal epithelial subtypes in the human small intestine. Many intestinal model systems include diverse intestinal epithelial subtypes, however, not in a physiologically relevant quantity or location and often lacking the progenitor stages of these subtypes. Our aim was to replicate a growth factor gradient in an induced pluripotent stem cell (iPSC)-derived Intestine-Chip, hereby maintaining proliferating stem cells and inducing the diverse stages of differentiating epithelial subtypes. Additionally, we characterise the intestinal mesenchymal population upon exposure to this gradient.
Methods. Human intestinal epithelial and mesenchymal cells were generated from three control iPSC lines, which were then introduced in an Emulate Intestine-Chip. The cells were exposed to 8expansion medium9 mimicking the condition in the crypt region, 8differentiation medium9 mimicking the condition in the villus region or a gradient by introducing these media to the lower and upper compartment of the system respectively. The intestinal epithelial and mesenchymal populations were assessed via immunefluorescent staining, flow cytometry and single-cell RNA sequencing. Barrier integrity was assessed using a FITC-Dextran 4kDa translocation assay.
Results. We could steer the intestinal epithelial subtype diversity by changing medium composition. The differentiation medium (in one or both compartments) increased the number of goblet cells, enteroendocrine cells, enterocytes, and Paneth cells, however, the proliferating transit-amplifying cells and tissue morphology were better preserved upon exposure to expansion medium basolaterally and differentiation medium apically. Moreover, the gradient resulted in both progenitor and mature stages of epithelial subtypes, while having differentiation medium in both compartments yielded mostly mature subtypes. The mesenchymal population drastically reduced upon exposure to differentiation medium and was enriched in fibroblast-like subtypes. Further analysis of the single-cell RNA sequencing dataset of the iPSC-derived Intestine-Chip will provide insight into the resemblance to reference data of the human intestine.
Conclusion. We present a thorough characterisation of the intestinal epithelial and mesenchymal populations in an iPSC-derived Intestine-Chip. By applying a growth factor gradient, we obtain a physiologically relevant intestinal epithelial composition, capturing the entire differentiation trajectory from stem cells to intermediate progenitor stages and mature cells.
Autologous co-cultures of human intestinal CD8+ cells and organoids on-chip to recapitulate a mucosal immune response
Joram Mooiweer1,2, Renée Moerkens1,2, Aarón D. Ramirez-Sanchez1, Gieneke Gonera1,3, Bana Jabri4,5, Robert Barrett6,7, Cisca Wijmenga1, Iris Jonkers1, Sebo Withoff1,2
1 Department of Genetics, University Medical Center Groningen, University of Groningen, Groningen, The Netherlands 2 Netherlands Organ-on-Chip Initiative (NOCI), Leiden, The Netherlands 3 Department of Paediatrics, Wilhelmina Hospital Assen, Assen, The Netherlands 4 Department of Medicine, University of Chicago, Chicago, IL, USA 5 Committee on Immunology, University of Chicago, Chicago, IL, USA 6 Cedars-Sinai Medical Center, Board of Governors Regenerative Medicine Institute, Los Angeles, CA, USA 7 Cedars-Sinai Medical Center, F. Widjaja Foundation Inflammatory Bowel and Immunobiology Research Institute, Los Angeles, CA, USA
Abstract
Coeliac Disease (CD) is a complex, multifactorial and immune-mediated disorder, characterised by a strong inflammatory response in the small intestine triggered by dietary gluten. The inflammation eventually results in the activation of CD8+ intraepithelial lymphocytes (IELs). The activated IELs attack and damage the intestinal epithelial cells (IECs), leading to villous atrophy. The molecular crosstalk between IELs and IECs driving the IEC destruction has however remained elusive. In this study we aim to elucidate these molecular drivers of villous atrophy in CD by establishing a novel patient-derived immunocompetent mucosal barrier-on-chip model of the small intestine.
For this, we generated human intestinal organoids (HIOs) from duodenal biopsies taken from CD patients and controls participating in the Coeliac Disease Northern Netherlands (CeDNN) biobank. In parallel, we were able to isolate and culture CD8³³+ TCR³³+ IELs from a duodenal biopsy of the same individual what allows for the generation of a temporary patient specific IEL cell line. These cells were used to set-up viable long-term autologous co-culture systems of IELs. With this, we can recapitulate the IEL migration towards, and interactions with IECs by life cell microscopy. CD relevant cytokines Interleukin 15 (IL-15) and IL-21 potentiate the IELs in co-culture, indicated by elevated Granzyme B (GzmB) levels. Also, functionality (i.e. the cytotoxic potential) of the IELs towards the HIOs can be followed as demonstrated by measuring apoptosis of the organoids.
This new autologous co-culture system thus allows for monitoring key aspects of the CD mucosal immune response. Currently we are performing side-by-side comparisons of patient-derived and control-derived co-cultures to identify differences in IEL activation and cytotoxic potential. We aim to do in depth characterisations of the IEL and IEC compartments separately using (single cell) RNA sequencing. This co-culture system will be further advanced by recreating the IEL-epithelial barrier interface in a microfluidic chip, which will provide a better controlled environment and allows integration of mechanical stimulation such as continuous fluid flow. Ultimately, this patient-derived model system will shed light on the molecular drivers of villous atrophy in CD and eventually will be used to test potential therapeutic interventions.
Intestinal organoids: a cellular model to study Coeliac Disease
M. Vittoria Barone, Claudia Bellomo, Francesca Furone, Merlin Nanayakkara
Department of translational Medicine (DISMET) and European Laboratory for the Investigation of Food Induced Diseases (ELFID) University Federico II, Naples, Italy
Abstract
Coeliac disease (CD] is an immune-mediated enteropathy triggered in genetically susceptible individuals by gluten containing cereals. It is characterised by gluten-induced symptoms, CD-associated autoantibodies, and an enteropathy. A central role in the pathogenesis of CD is played by the HLA-restricted gliadin-specific intestinal T cell response generated in a pro-inflammatory environment [1]. How is this pro inflammatory environment generated is still not clear. In vivo studies showed on a population at risk, before the onset of the disease and, interestingly, before the introduction of gluten in the diet, cellular and metabolic alterations in absence of the T-cell mediated response [2].
Intestinal organoids are defined as self-organizing three- dimensional structures that closely mimic the in vivo situation, they can be generated from adult stem cells, located in the crypts of the intestinal epithelium [3]. We have generated intestinal organoids from CD patients at different stage of the diseases to understand the role of the intestinal epithelium in the CD intestinal lesion.
We have shown from in vitro study on CD biopsies and organoids that a constant low-grade inflammation is present in epithelial cells also in absence of gluten. In fact, inflammation was present not only in CD biopsies in the acute and remission phase of the disease, but also in Potential patients9 biopsies before the onset of the intestinal lesion. this inflammation was reproduced also in intestinal organoids derived from CD patients and contrary to IBD patients9 organoids it was persistent after several passages in culture [4]. Interestingly cytokinoma analysis of the CD organoids showed 25 out of 27 pro-inflammatory cytokines increased. Moreover, the supernatant of CD organoids was able to induce inflammation of controls organoids. Intestinal organoids can be a good model to study inflammation in CD patients and to test pro-pre-post-biotics or nutraceuticals that could prevent the epithelial inflammation [5-8].
Inflammation, probably constitutive, could have a main role in CD. Adding this disease <tout court= to the big family of increasing chronic inflammatory diseases where nutrients can have pro-inflammatory or anti-inflammatory effects, directly or indirectly mediated by the intestinal microbiota, where the intestine and in particular the intestinal epithelial cells, can function as a crossroad for the control of the inflammation both local and at distance.
References
1. Sollid LM, Jabri B. Triggers and drivers of autoimmunity: lessons from coeliac disease. Nat Rev Immunol 2013; 13(4): 660 294-302.
2. Barone MV, et al. Celiac Disease, an example of chronic inflammatory disease with fragile intestine. IJMS 2022; 23:71.
3. Perrone F, Zilbauer M. Biobanking of human gut organoids for translational research. Exp Mol Med 2021; 53:145131458.
4. Porpora M, et al. Inflammation Is Present, Persistent and More Sensitive to Proinflammatory Triggers in Celiac Disease Enterocytes. Int J Mol Sci 2022; 23: 1973.
5. Conte M, et al. Gliadin Peptide P31-43 Induces mTOR/NFk³ Activation and Reduces Autophagy: The Role of Lactobacillus paracasei CBA L74 Postbiotc. Int J Mol Sci 2022; 23: 3655.
6. Furone F, et al. The protective role of Lactobacillus rhamnosus GG postbiotic on the alteration of autophagy and inflammation pathways induced by gliadin in intestinal models. Front Med (Lausanne) 2023; 10: 1085578.
7. Labruna G, et al. Celiac disease-associated Neisseria flavescens decreases mitochondrial respiration in CaCo-2 epithelial cells: Impact of Lactobacillus paracasei CBA L74 on bacterial-induced cellular imbalance. Cell Microbiol 2019; 21: e13035.
Organoids in the study of coeliac pathogenesis
Violaine Dony, Michael Schumann
Gastroenterology, Charité 3 Universitätsmedizin Berlin, Berlin, Germany
Introduction
Meanwhile, organoids are a well-established tool in gastrointestinal research to study the epithelial contribution to mucosal diseases as coeliac disease (CD), inflammatory bowel diseases or cancer. When isolated from endoscopic or surgical mucosa-samples, they carry as primary epithelial cell cultures the epithelial properties of the human individual they were collected from 3 including the disease determining alterations. Intestinal organoids are kept in culture in their original 3D confirmation, growing in basement membrane-like matrices in media that are supplemental with a distinct combination of growth factors including Wnt, Rspondin, Noggin, and Epidermal Growth Factor (EGF) that are selected depending on the cell differentiation the experiment is designed for. For the purpose of physiological studies of barrier function or mucosal substance transport, they can be re-seeded on coated filter supports to yield a 2D organoid culture, which can be used for measurements of transepithelial resistance or macromolecular permeability.
In coeliac research, the search for additional factors triggering a disease flare is ongoing. GI infections or mere changes of microbial colonisation of the intestinal mucosa are considered to trigger a flare and would thereby also explain why the disease 3 in a significant proportion of patients 3 develops only later in life. Such infections or changes in microbiota composition might trigger subtle barrier defects and might thereby contribute to the induction of CD.
Thus, our goal is to study the capacity of microorganisms in altering epithelial cell polarity and barrier function in intestinal epithelial cells and to determine if microbiota changes can contribute to increasing the transport of gluten peptides across the epithelia.
Methods
The study was structured in two phases. In phase I (screening), cell lines including Caco2-Bbe and Caco2-PIP2, transfected with a GFP-tagged PIP2 binding moiety to analyse cell polarity, were exposed to various microorganisms. Selection of microorganisms for phase II was based on four criteria: decrease of transepithelial resistance (TER), impact on tight junction (TJ) and junctional / polarity proteins in confocal microscopy and increase of paracellular permeability using sandwich assays.
In phase II, selected microorganisms were added to the apical compartment of small intestinal 2D organoids. Barrier function regarding small solutes and macromolecules (including the gliadin fragment 33mer) and cell polarity was determined.
Results
Salmonella typhimurium, Pseudomonas aeruginosa, uropathogenic Escherichia coli, Escherichia coli K12 and Candida albicans, Saccharomyces cervisiae were selected as microorganisms for Phase I. TER measurements, ZO1 immunostainings and macromolecular permeability assays (TMR-dextran3000) revealed a barrier defect in Salmonella typhimurium, Pseudomonas aeruginosa, uropathogenic Escherichia coli and Candida albicans, but not in Escherichia coli K12 and Saccharomyces cervisiae. Altered epithelial polarity as uncovered by the PIP2 tracker was specifically detected in Pseudomonas-exposed organoid monolayers. For phase II, P. aeruginosa and C. albicans were selected. Exposure to duodenal organoids revealed comparable results to the Caco-cell systems (Fig. 1: TER; Fig. 2: ZO1 staining). After Pseudomonas exposure, polarity was severely altered as uncovered by Ezrin staining. Gliadin translocation was determined by the sandwich assay using a biotin- and TMR-tagged 33mer-gliadin fragment and was increased after C. albicans exposure (Fig. 3).
Summary/Conclusion
These results indicate that Pseudomonas and Candida have the capacity to induce an alteration of epithelial polarity / barrier change in epithelial monolayers. Candida also contributes to 33mer-uptake. Candida-induced effects on barrier function were dependent on its epithelial-invasive property.
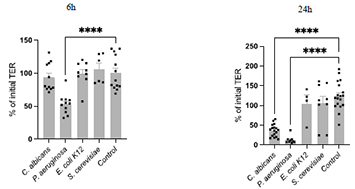
Figure 1. Transepithelial resistance (TER) in 2D organoids of a healthy control patient after exposure to the indicated microbiota.
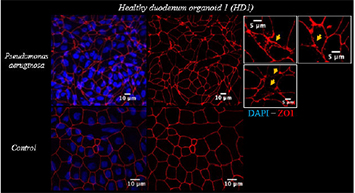
Figure 2. Immunostaining of ZO1 reveals the extent of morphological tight junction integrity in duodenal 2D organoids of a healthy individual.
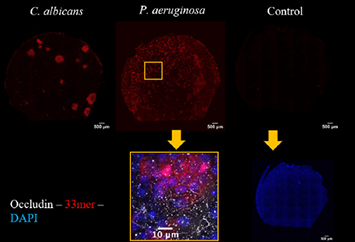
Figure 3. Gliadin-33mer translocation as measured by sandwich assay in duodenal 2D organoids of a coeliac individual after microbiota exposure.
Statements on current developments concerning gluten analysis, clinical and legal aspects
Update on Codex issues regarding gluten
Hertha Deutsch
AOECS Codex Delegate, Austrian Coeliac Society, Vienna
Abstract
AOECS, the Association of European Coeliac Societies, has Observer status in the Codex Alimentarius Commission and the Committees since 1992 and worked successfully to improve Codex Standards for the benefit of coeliacs. In 1999, the Codex Alimentarius Commission adopted the list of foods and ingredients which are known to cause hypersensitivity and shall always be declared. The first on the list are <Cereals containing gluten, i.e. wheat, rye, barley, oats, spelt or their hybridised strains and products of these=.
Proposed Draft Revision of the General Standard for the Labelling of Prepackaged Foods 3 Provisions relevant to Allergen Labelling
In October 2021, the Codex Committee on Food Labelling (CCFL) discussed provisions relevant to allergen labelling and continued in the next session in May 2023. AOECS participated in the sessions, in pre-meetings and elaborated several comments, which were distributed to all CCFL participants. We worked successfully to delete coeliac disease from the definition of <Food allergy=, to insert a text of coeliac disease in the definition of terms and that the definition of <Allergen= covers also <other specific immune-mediated reactions= and not only IgE-mediated reactions. However, further efforts are requested to improve the proposed texts e.g. that the definition of gluten should be in accordance with the Codex Standard for Foods for Special Dietary Use for Persons Intolerant to Gluten (CXS 118-1979), to solve the confusion regarding the requested <Specified Name= to label <wheat= when the ingredient is spelt or a gluten-free wheat starch or an ingredient derived from wheat which does not contain gluten, and also regarding oats. CCFL agreed to forward the proposed Draft Revision of the General Standard for the Labelling of Prepackaged Foods - Provisions relevant to Allergen Labelling to the Codex Alimentarius Commission for adoption at Step 5, Step 8 is the final adoption.
Proposed Draft Guidance on Precautionary Allergen Labelling (PAL)
In May 2023, the CCFL discussed the PAL and AOECS did not agree to the draft because coeliac disease is not considered. The proposed RfD of 5 mg wheat does not match with the threshold of gluten-free <20 mg/kg gluten as defined in CXS 118-1979. CCFL requested the Codex Committee on Methods of Analysis and Sampling (CCMAS) to recommend suitable analytical methods and guidance on their validation and applications. At the CCMAS session, AOECS informed that the definition of food allergen agreed by CCFL includes also other specific immune-mediated reactions, which is coeliac disease, and requested that the CXS 118-1979 should be taken into account in the answer to CCFL. CCMAS was not ready to provide a reply to CCFL at this time and agreed to establish an EWG to work on it.
The reports of the CCFL and CCMAS are published on the Codex website www.fao.org.
Prolamin Working Group: Challenges of the next years from the industry's perspective
Götz Kröner
Kröner-Stärke GmbH, Ibbenbüren, Germany
Abstract
Challenges of the next years regarding gluten-free foods from the industry9s perspective.
1. Existing maximum residue level of 20 ppm gluten for gluten-free foods Concerns of coeliac patients whether the limit is low enough.
Action needed:
• Empiric study to investigate the suitability of the limit of 20 ppm gluten in reality.
• How did nutrition habits change since the limit of 20 ppm gluten entered into force?
• Is there an impact on the personal feeling of the affected people?
2. Acceptance of gluten-free wheat starch in baby food Despite better functionality baby food producers reject using gluten-free wheat starch due to missing acceptance by the consumer.
Action needed:
• Scientifically sound information for the consumer must be provided.
3. Existing maximum residue level of 20 ppm gluten in gluten-free wheat starch Quest for lower maximum residue level of 5 ppm gluten in gluten-free wheat starch.
Action needed:
• Investigation of the allergenic potential of proteases which are necessary to obtain residual gluten contents of 5 ppm or less.
• Comparison with <naturally= processed wheat starch complying with the 20 ppm limit for gluten.
4. Wheat starch in packaging materials for food Concerns of migration of gluten from the packaging material into the food.
Action needed:
• Scientifically sound data must be provided.
Perspectives and action plan of the PWG
Peter Koehler1, Carmen Gianfrani2
1 Biotask AG, Esslingen, Germany 2 Institute of Biochemistry and Cell Biology, CNR, Naples, Italy
The Prolamin Working Group executive meeting and joint discussion held on 22 September 2023, led to the decisions and statements outlined below.
Action plan
I. Analytical
• The PWG gliadin reference material is available from Arbeitsgemeinschaft Getreideforschung e. V. (Association of Cereal Research), Schuetzenberg 10, 32756 Detmold, Germany, E-mail: info@agf-detmold.de.
• The price for one batch (100 mg) is 150 Euro.
• Material for at least 4 years is still on stock.
• The future of PWG gliadin is unclear. Plans to prepare new PWG gliadin reference material must be reconsidered by the group!
II. Clinical
An international consortium of PWG-members and associated partners has successfully applied for an EU grant running from 2022 to 2025. The ImmunoSafe-CeD project aims to determine the CD immunogenic activity of intact and partially hydrolysed gluten from wheat, rye and barley and to develop improved comprehensive functional and analytical assays. Supervisor: Katharina Scherf.
III. Members, Policy
Carmen Gianfrani was elected as new chairperson of the group. Knut Lundin is acting as the deputy chairperson.
• Peter Koehler has resigned as chairperson and has left the group.
• Iris Jonkers from University of Groningen, The Netherlands, is a new member of the group.
• The International Society for the Study of Celiac Disease (ISSCD) has contacted PWG to discuss about how the two organisations can interact. The contact person is Iris Jonkers.
• A new mission statement of the group will be worked out and will be published on the PWG homepage.
• Proceedings of this meeting will be available free of charge in electronic form from the PWG website (http://www.wgpat.com).
Next meeting: 2024
We are very pleased to announce the venue for our meeting:
Darmstadt, Germany
Hosts:
R-Biopharm AG
Registration office: German Coeliac Society (DZG)
Time: 26. 3 28. September 2024
Focus of the meeting: To be announced
The meeting will be limited to 55 participants and attendance is by invitation only.
Invitations will be sent by April 2024. Registration deadline will be May 12, 2024.
Very special thanks to the hosts for this kind invitation!
|
|
List of Participants
GROUP MEMBERS
Prof. Dr. Carlo Catassi
Università Politecnica delle Marche
Department of Pediatrics
Via Corridoni 11
60123 ANCONA, ITALY
Phone: +39 071 5962364
E-mail: c.catassi@staff.univpm.it
Prof. Dr. Fernando G. Chirdo
Universidad Nacional de La Plata
Facultad de Ciencias Exactas
Instituto de Estudios Immunologicos y
Fisiopatologicos - IIFP
Calle 47 y 115
1900 LA PLATA, ARGENTINA
Phone: +54 221 423 5 333 (Int 45)
E-mail: fchirdo@biol.unlp.edu.ar
Prof. Dr. Paul J. Ciclitira
University of East Anglia
Medical School
Bob Champion Building
James Watson Road
BR4 7UJ NORWICH
UNITED KINGDOM
Phone: +44 203 751 1104
E-mail: pciclitira@btinternet.com
Prof. Dr. Conleth Feighery
University of Dublin, Department of
Immunology, St. James’s Hospital
James’s Street
DUBLIN 8, IRELAND
Phone: +353 879969041
E-mail: cfighery@tcd.ie
Dr. Carmen Gianfrani
Institute of Biochemistry and
Cell Biology - CNR
Via Pietro Castellino 111
80131 NAPLES, ITALY
Phone: +39 081 6132224
E-mail: c.gianfrani@ibp.cnr.it
Prof. Dr. Peter Koehler
Biotask AG
Schelztorstraße 54-56
73728 ESSLINGEN, GERMANY
Phone: +49 711 31059068
E-mail: peter.koehler@biotask.de
Prof. Dr. Frits Koning
Leiden University Medical Centre, E3-Q
Department of Immunohaematology
and Bloodbank
Albinusdreef 2
2333 ZA LEIDEN, THE NETHERLANDS
Phone: +31 715 266673
E-mail: fkoning@lumc.nl
Prof. Dr. Knut Lundin
University of Oslo
Institute of Clinical Medicine
Postboks 1171, Blindern
0881 OSLO, NORWAY
Phone: +47 90980325
E-mail: knut.lundin@medisin.uio.no
Prof. Dr. Stefania Masci
University of Tuscia
Department of Agricultural and Forest
Sciences (DAFNE)
Via San Camillo de Lellis s.n.c.
01100 VITERBO, ITALY
E-mail: masci@unitus.it
Prof. Dr. Katharina Scherf
Karlsruhe Institute of Technology (KIT)
Institute of Applied Biosciences
Department of Bioactive and
Functional Food Chemistry
Adenauerring 20 a
76131 KARLSRUHE, GERMANY
Phone: +49 721 608 42929
E-mail: katharina.scherf@kit.edu
Prof. Dr. Dr. Detlef Schuppan
I. Medizinische Klinik und Poliklinik
Universitätsmedizin der Johannes
Gutenberg-Universität Mainz
Institut für Translationale Medizin
Langenbeckstraße 1
55131 MAINZ, GERMANY
Phone: +49 6131 177355/177356/177104
E-mail:
detlef.schuppan@unimedizin-mainz.de
Dr. René Smulders
Wageningen University & Research,
Plant Research
Droevendaalsesteeg 1
6708 PB WAGENINGEN,
THE NETHETRLANDS
Phone: +31 620298266
E-mail: rene.smulders@wur.nl
Prof. Dr. Riccardo Troncone
University Federico II
Department of Pediatrics
Via Pansini 5
80131 NAPLES, ITALY
Phone: +39 3483132274
E-mail: troncone@unina.it
HOSTS
Dr. René Smulders
Wageningen University & Research,
Plant Research
Droevendaalsesteeg 1
6708 PB WAGENINGEN,
THE NETHETRLANDS
Phone: +31 620298266
E-mail: rene.smulders@wur.nl
Dr. Ingrid van der Meer
Wageningen University & Research
Bioscience Droevendaalsesteeg 1 6708 PB
WAGENINGEN, THE NETHETRLANDS
E-mail: ingrid.vandermeer@wur.nl
Dr. Peter Weegels Sonneveld
Group BV Rietgorsweg 1 3356 LJ
PAPENDRECHT, THE NETHERLANDS
E-mail: peter.weegels@sonneveld.com
Ms. Daniëlle van der Wee-Uittenbogaard
Wageningen University & Research
Secretriaat Plant Breeding Droevendaalsesteeg 1 6708 PB
WAGENINGEN, THE NETHETRLANDS
E-mail: danielle.vanderwee@wur.nl
INVITED SPEAKERS
Dr. Iris Jonkers
University of Groningen
Faculty of Medical Sciences (UMCG) Antonius Deusinglaan 1, 9713 AV
GRONINGEN THE NETHERLANDS
E-mail: i.h.jonkers@umcg.nl
Mr. Joram Mooiweer
University of Groningen
Faculty of Medical Sciences (UMCG) Antonius Deusinglaan 1, 9713 AV
GRONINGEN THE NETHERLANDS
E-mail: j.mooiweer@umcg.nl
Prof. Maria Vittoria Barone
University of Naples Federico II Department of Translational
Medical Sciences, Via Sergio Pansini 5 80131
NAPLES, ITALY
E-mail: mariavittoria.barone@unina.it
Dr. Michael Schumann
Charité - Universitätsmedizin Berlin Hindenburgdamm 30 12200
Berlin, Germany
E-mail: michael.schumann@charite.de
GUESTS
Ms. Tova Almlöf
Semper AB Box 1101 SE 17222
SUNDBYBERG, SWEDEN
E-mail: tova.almlof@semper.se
Ms. Sofia Beisel
Deutsche Zöliakiegesellschaft e.V. Kupferstraße 36 70565
STUTTGART, GERMANY
E-mail: sofia.beisel@dzg-online.de
Dr. Markus Brandt
Ernst Böcker GmbH & Co KG Ringstrasse 55-57 32427
MINDEN, GERMANY
E-mail: markus.brandt@sauerteig.de
Dr. Gaetano Cardone
Dr. Schaer S.p.A. Winkelau 9 39014
BURGSTALL, ITALY
E-mail: gaetano.cardone@drschaer.com
Dr. Johan De Meester
Cargill R&D Centre Europe Havenstraat 84 B-1800
VILVOORDE, BELGIUM
E-mail: Johan_De_Meester@cargill.com
Ms. Hertha Deutsch
Österreichische Arbeitsgemeinschaft Zöliakie Anton Baumgartner Straße 44/C5/2302 1230
VIENNA, AUSTRIA
E-mail: hertha.deutsch@chello.at
Dr. Mark Driscoll
Neogen Europe The Dairy School, Auchincruive KA6 5 HU
AYR, UNITED KINGDOM
E-mail: mdriscoll@neogen.com
Dr. Margareta Elding-Pontén
Fria Bröd AB Gröenvägen 2 43891
LANDVETTER, SWEDEN
E-mail: Margareta.Elding-Ponten@fria.se
Ms. Maren Finke
Hermann Kröner GmbH Lengericher Straße 158 49479
IBBENBÜREN, GERMANY
E-mail: finke@kroener-staerke.de
Dr. Carlos Galera
Hygiena Diagnostica Espana P.I. Parque Plata Calle Cañada Real 31-35 41900 CAMAS,
SEVILLA, SPAIN
E-mail: cgalera@hygiena.com
Ms. Emily Hampton
Coeliac UK 3rd Floor, Apollo Centre Desborough Road HP11 2QW HIGH
WYCOMBE, UNITED KINGDOM
E-mail: Emily.Hampton@coeliac.org.uk
Dr. Xin Huang
University of Helsinki Natural Resources Institute Finland Latokartaninkaari 9 790
HELSINKI, FINLAND
E-mail: xin.huang@helsinki.fi
Dr. Götz Kröner
Hermann Kröner GmbH Lengericher Str. 158 49479
IBBENBÜREN, GERMANY
E-mail: kroener@kroener-staerke.de
Ms. Manjusha Neerukonda
University Medical Center Mainz Langenbeckstraße 1 55131
MAINZ, GERMANY
E-mail: manjusha@uni-mainz.de
Dr. Luisa Novellino
Associazione Italiana Celiachia APS Via Caffaro, 10 16124
GENOVA, ITALY
Email: l.novellino@celiachia.it
Mr. Cristóbal Pérez Sixto
Associació Celíacs de Catalunya St/Independencia 257,
ground floor 8026
BARCELONA, SPAIN
E-mail: cristobal@celiacscatalunya.org
Ms. Stine Rosenqvist Lund
Oslo University Hospital (OUS) Sognsvannsveien 20 372
OSLO, NORWAY
E-mail: strolu@ous-hf.no
Mr. Stefan Schmidt
R-Biopharm AG An der neuen Bergstraße 17 64297
DARMSTADT, GERMANY
Phone: +49 151 29808524
E-mail: st.schmidt@r-biopharm.de
Dr. Juan Ignacio Serrano-Vela
Coeliac Disease & Gluten Sensitivity Association Madrid Calle Lanuza 19-bajo 28028
MADRID, SPAIN E-mail: nachoserrano@celiacosmadrid.org
Ms. Eline Smits
University of Groningen Faculty of Medical Sciences (UMCG) Antonius Deusinglaan 1, 9713 AV
GRONINGEN THE NETHERLANDS
E-mail: e.smits@umcg.nl
Dr. Karoline Terberger
Böcker Sauerteig GmbH & Co. KG Ringstraße 55-57 32427
MINDEN, GERMANY
E-mail: karoline.terberger@sauerteig.de
Dr. Niklas Weber
R-Biopharm AG An der neuen Bergstraße 17 64297
DARMSTADT, GERMANY
E-mail: n.weber@r-biopharm.de
Dr. Thomas Weiss
R-Biopharm AG An der neuen Bergstraße 17 64297
DARMSTADT, GERMANY
E-mail: t.weiss@r-biopharm.de
Impressum
Proceedings of the 36th Meeting
WORKING GROUP
on PROLAMIN ANALYSIS and TOXICITY
21 3 23 September 2023 Wageningen, The Netherlands
This work including all parts is subject to copyright. All rights are reserved and any
utilisation is only permitted under the provisions of the German Copyright Law.
Permissions for use must always be obtained from the publisher. This is in particular
valid for reproduction, translation, conversion to microfilm and for storage or
processing in electronic systems.
Scientific Organisation
Prof. Dr. Peter Koehler
biotask AG Schelztorstraße 54-56, 73728
ESSLINGEN, GERMANY
Phone: +49 711 31059068; Fax: +49 711 31059070
E-mail: peter.koehler@biotask.de
Hosts
René Smulders, Twan America, Ingrid van der Meer, Peter Weegels
Wageningen University & Research, Droevendaalsesteeg 1, 6708 PB
Wageningen, THE NETHERLANDS
Phone: +31 6 20298266
E-mail: rene.smulders@wur.nl
Cover picture* and picture of participants
Peter Koehler
over picture: Small lily pond in the Arboretum De Dreijen, Wageningen. In the centre part of the picture, surrounded by trees, a sculpture of Carl von Linné is located.
© Carmen Gianfrani and Peter Koehler 2023
|


