Proceedings of the 32nd Meeting
Working Group on Prolamin Analysis
and Toxicity (PWG)
Edited by Peter Koehler
Esslingen, March 2019
Preface
Already in the 2015 meeting of the Working Group on Prolamin Analysis and Toxicity
(PWG) in Tulln, Austria, Pauline Titchener from Neogen Europe Ltd. informed me
that Neogen would be willing to host one of the following PWG meetings in their
European headquarter in Scotland. Finally, 2018 was the year that this became reality.
The meeting was held at Oswald Hall, a historic building belonging to the Neogen
campus in Ayrshire. The lecture room was very special with its historic interior
decoration but also with modern facilities that are needed for a conference. Pauline and
her team were present during the entire meeting. Pauline was also available after the
official programme and organised a joint whisky tasting and dinner. Apart from the
group members the audience comprised an invited speaker, guests from academia,
industry, and international coeliac societies. Representatives from cereal starch
producers, producers of gluten-free foods, as well as manufacturers of kits for gluten
analysis and of kits for antibody tests in the serology of coeliac disease (CD)
participated from industry.
Analytical and clinical work in the field of CD, non-coeliac gluten/wheat sensitivity
(NCGS/NCWS), wheat allergy and gluten done in the labs of PWG members as well
as results of guests and invited speakers were presented in 18 talks and lively
discussed at the meeting. This time, legal and regulatory aspects of gluten analysis
were not discussed because only a few participants from coeliac societies were able to
attend the meeting. A symposium with presentations looking at the successful
determination of the wheat genome and its exploitation in future research was
organised and highly estimated by the audience.
I am grateful to all participants for their active contributions as presenters as well as
during the discussions. This made the 2018 meeting a great success. I would like to
express my special thanks to Neogen Europe Ltd., in particular to Pauline Titchener,
for being a perfect host as well as to Sharon Forsythe for her help in the organisation
of the meeting. Special thanks go to Thomas Mothes and Martin Stern for their longtime
dedication to the group. Both left the group by the end of 2018. Finally, I express
my gratitude to all friends, colleagues, sponsors and participants for their inspiration
and continuing support of the PWG.
Esslingen, March 2019, Peter Koehler
Executive Summary
Eighteen presentations covered all aspects related to gluten, coeliac disease (CD) and
other relevant hypersensitivities. Sixteen authors have sent manuscripts that are
compiled in this proceedings book. Some coeliac societies were not able to send
delegates because of a meeting of the Association of European Coeliac Societies
(AOECS) at the same time. Therefore, legal aspects were not covered in this meeting.
Analytical session
Seven presentations covered the analysis of gluten and other proteins of interest for the
PWG. It became obvious that ELISA is currently the method of choice for gluten
quantitation because this method was used in five presentations. Apart from the
comparison of G12 and R5 ELISAs, a new ELISA for total gluten using three
monoclonal antibodies was introduced. The unsolved problem of suitable reference
materials for gluten quantitation was covered in two talks. Finally, two studies were
presented that used LC-MS. One presentation provided data on the concentrations of
amylase-trypsin inhibitors (ATI) in different wheat species, and in another study,
isopeptides between tissue transglutaminase and wheat gluten peptides were identified.
Clinical session
One of the six presentations provided evidence for reduced activity of Einkorn in CD
because of low resistance of peptides to proteolytic cleavage. Another study showed
that there appears to be a discrepancy between serology and inflammation in potential
CD. The third presentation showed that there are several pathways of apoptosis in
untreated CD. Studies on the kinetics and transcriptomic profile of antigen-specific
cells after gluten challenge showed that sampling between day 6 and 8 after 3-day
gluten challenge is an appropriate time window for collection of gluten-specific T
cells. Finally, there is evidence that deamidated gliadins worsen immune reactions in
wheat allergy. This is of practical relevance because some industrial processes use
deamidation to functionalise gluten proteins.
Symposium: Wheat genomics
The symposium included two presentations of recognised experts in wheat genomics.
In a very exciting talk, the latest results of genome sequencing were reported. These
activities resulted in the first wheat reference genome, and it was discussed, how this
novel knowledge can be exploited in the near future. The second presentation was also
very interesting and dealt with the latest approaches on wheat genome editing by the
CRISPR/Cas technology. The first results of a study on eliminating coeliac active
epitopes in wheat were discussed. The symposium showed that genome editing and
sequencing are currently among the leading scientific topics in cereal research.
Analytical research reports
AOAC International update: Gluten in oats method
validation framework
Paul Wehling, Harrison Feldkamp
General Mills, Inc., Minneapolis, MN, USA
Introduction
In September 2017, the AOAC International Stakeholder Panel for Alternative
Methods (ISPAM) adopted Standard Method Performance Requirement (SMPR)
2017.021, Quantitation of Wheat, Rye, and Barley Gluten in Oats, as the guidance
document for the validation of methods for measuring gluten in oat products [1].
In the past, gluten methods were evaluated for accuracy based on spiking wheat gluten
into various gluten-free (GF) matrices and estimating recovery of the method by
calculating the percentage of analyte recovered during a multi-lab collaborative study.
Recent quantitative methods, such as AOAC OMA methods 2012.01 and 2014.03
have used this process. In the case of validating an ELISA method for gluten in oats, it
will be essential to evaluate the kit responses to not only wheat, but also barley and
rye. The SMPR 2017.021 has indicated that for this method project, the responses of
wheat, rye and barley should be estimated independently as part of single-lab
validation. In order to facilitate such validations, a series of samples were prepared,
each spiked with a single grain at specific levels. The SMPR states that for approval as
an OMA method, the candidate method must demonstrate recovery of wheat, rye and
barley gluten proteins separately, and the recoveries must be between 50% and 200%.
This represents a new approach to the validation of gluten methods, where historically
only wheat proteins have been considered as relevant to method accuracy.
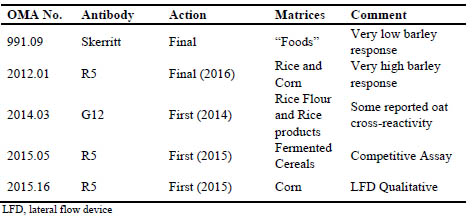
Tab. 1 shows current AOAC Official Methods of Analysis (OMA) which have been
validated for analysis of gluten in foods. Recent practice within AOAC is to restrict
approval of the method to matrices which were studied in single lab validation (SLV)
and/or in multi-lab validations (MLV), such as with a collaborative study. There
currently is no OMA method applicable to oat products. Oat products are unique in the gluten-free supply chain in that there is a significant probability of encountering lowlevel
barley contamination from agricultural commingling due to geographical areas
where oats and barley are grown contiguously, specifically in Northern United States,
and Western Canada. In the past 10 years, oat processors have developed systems to
produce GF oats, either by mechanical/optical separation, or by selective growing/IP
agricultural processes. In both of these systems, barley and wheat contamination are
the most common sources of gluten containing grains. As such, it is critical that a method being used to inspect and control a GF oat process be accurate to both wheat
and barley proteins.
In the past, most of the focus of calibrating and validating gluten methods was on
accuracy to wheat proteins. The Skerritt antibody was developed originally to measure
wheat and has very low response to barley proteins [2]. The R5 antibody was raised
against rye proteins and has reported high response to barley proteins [3]. Both
methods were successfully validated with acceptable recovery of wheat gluten
proteins.
Table 1. Current AOAC International Official Methods of Analysis (OMA) for gluten
Materials and methods
In order to validate the response of a method to wheat, rye and barley proteins
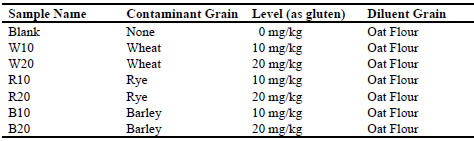
separately, AOAC has produced a series of reference materials. This series consists of
seven spiked samples, which are made from GF oat flour, quantitatively spiked with
various levels of wheat, rye and barley flours. Tab. 2 shows the spike levels of each of
the seven materials.
Table 2. AOAC International reference samples for gluten validation
Spiking materials
GF Oats were obtained from General Mills, Inc., USA, by optical and mechanical
sorting, dehulling and further optical sorting of dehulled groats. Oat groats were then
milled with a Retsch Mill ZM200 to obtain oat flour, which tested at <1 mg/kg by the
R5 method by replicate analysis (mean of 18 reps at 5 g test portion).
For rye and barley spike materials, blends were made for each grain from several
samples of selected grain cultivars obtained from seed breeders in the region. In the
case of rye, eight separate cultivars were blended in equal parts, then milled to flour to
obtain the blended spike material. For barley, six cultivars of 2-row barley, plus three
cultivars of 6-row barley were milled, then blended together to obtain a spiking flour.
For wheat, we were unable to obtain pure cultivars, so we instead used a mixture of
commercially available whole-wheat flours, and flours made from commercially
obtained wheat samples. In all, ten samples of wheats and whole wheat flours were
blended to make a spike flour representative of North American wheats grown in
2015-2017.
Characterisation of Spiking Materials
The three spiking flour blends were analysed for total protein by Dumas (N x 5.83)
nitrogen method. In order to estimate the level of gluten in each of the three spiking
blends, the AOAC Working Group approved the use of a wet chemical extraction
method to extract off non-gluten proteins and analyse the remaining solid pellet by
Dumas nitrogen and compare to the unextracted protein level. This extraction method
was based on the Codex Alimentarius definition of gluten as “the protein fraction from
wheat, rye or barley to which some persons are intolerant and that is insoluble in water
and 0.5 M NaCl.” [4]. The following method was used to estimate gluten levels in the
spiking materials.
1. Mill the grains through Retsch Mill ZM 200 with 0.5mm screen.
2. Weigh 150 mg sample grain into a 2 mL microcentrifuge tube. Record the weight to the
nearest 0.1 mg.
3. Add 1.5 mL water to the tube. Cap and vortex to completely disperse the sample.
4. Let the sample stand at ambient temp for 15 min, vortexing every 5 min.
5. Centrifuge in micro centrifuge for 10 min at 3400 RPM.
6. Decant off the supernatant, making sure not to lose any solids. If solids are not
completely at bottom of the tube, recentrifuge an additional 10 min.
7. Repeat steps 3-6 with water.
8. Repeat steps 3-6, 2 times with 0.5 M NaCl/PBS solution.
9. Place the tube in vacuum oven and dry overnight at 70 C under vacuum for 16 hours.
10. Remove from vacuum oven, put pellet in Dumas foil and drop in furnace to measure
nitrogen content. Use original flour weight as mass for Dumas calculation
11. Report N2 content per sample weight of original sample before washing.
12. Compare N2 content vs Dumas reading with no solvent treatment.
13. If needed, report % protein as %N x 5.83
Results and discussion
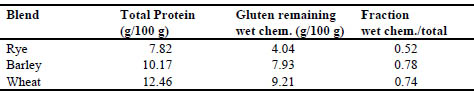
The three samples of flour spiking materials were analysed five times by Dumas
protein and five times by the wet chemical method above. Tab. 3 gives results of the
characterisation.
Table 3. Observed gluten levels of spiking materials
The dilution and manufacturing of the reference materials was performed by Trilogy
Labs, Washington, MO, USA to produce the series of seven samples as given in Tab.
2. The Materials are available for purchase through United States Pharmacopeia,
Rockville MD, USA, (Cat. No. 1294839).
As a demonstration of the suitability of the materials and in order to provide an
example of the process for estimating recovery, we have analysed each of the seven
samples with replication (18 replicates at 5 g test portion level) by the R5 method (RBiopharm
kit R7001) and report the results as follows in Tab. 4. Figure 1 is a plot of
these data.
Table 4. Observed gluten levels of reference materials by R5 antibody
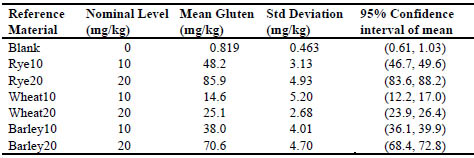
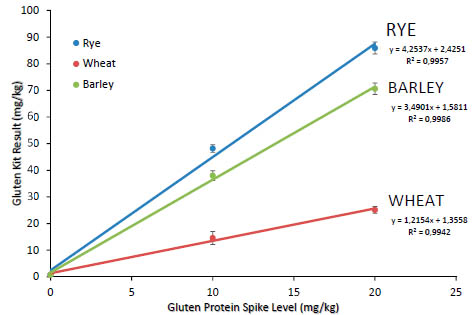
Figure 1. Plot of response for R5 kit to spiked recovery samples
To estimate overall recovery, we recommend plotting all individual replicates as
observed results vs. spiked nominal value and regressing as linear model ordinary
least-squares regression. The slope of the regression line will be the recovery estimate.
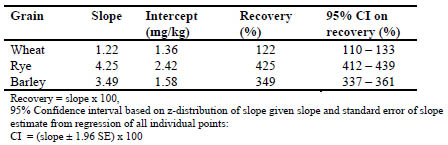
Tab. 5 is a summary of the recovery data obtained for the three grains.
Table 5. Statistical summary of R5 kit response plot
Conclusions
This new AOAC protocol represents a new approach to validating gluten methods.
While in the past, emphasis was put on accuracy with respect to wheat proteins, here
we are looking as well at trying to achieve balanced response by all three possible
gluten sources.
The AOAC SMPR requires a proposed method to demonstrate recoveries on all three
grains between 50 and 200%. By the analysis shown here, the R5 antibodies, in
conjunction with the current Mendez Cocktail extraction (AOAC OMA Method
2012.01) would not be considered suitable for use in oat products due to its high
responses for barley and rye (349% and 425% recovery, respectively). This overresponse
is higher than reported in prior reports [3] where the recovery was estimated
to be around 200% with the PWG gliadin calibrators and 2 x gliadin correction factor.
The estimates in that reference were made based on extracted proteins in solution, not
against grains spiked into grains, then extracted by a set method protocol. We feel the
results given here are better estimates for the recovery of the complete method,
including extraction. It is very important when estimating method recovery to include
the extraction steps from the method under study in the experiment. We have observed
very different recoveries on these reference materials with different extraction
methods, even with the same antibody system.
In addition, it should be noted that these reference materials are spiked samples, and
the contaminant grains have been milled prior to spiking. This makes the repeatability
of the methods very tight as opposed to incurred samples, where the particle sizes of
the contaminant grains are larger. We recommend that these reference materials be
used only for recovery studies, and not for precision estimates, as the precision
estimates on these reference materials will be much lower than observed on naturally
incurred oat flour samples. Precision studies should only be carried out with incurred
samples of the specific matrices under study.
References
1. 1. Boison J, Allred L, Almy D, et al, AOAC Standard Method Performance
Requirement 2017.021, J AOAC Int 2018; 101 (4): 1238-1242.
2. Skerritt J H, Hill A S. Enzyme immunoassay for determination of gluten in foods:
Collaborative study. J AOAC Int 1991;74:257-264.
3. Valdés I, Garcia E, Llorente M, Méndez E. Innovative approach to low-level gluten
determination using a novel sandwich enzyme-linked immunosorbent assay
protocol. Eur J Gastroenterol Hepatol 2003;15: 465-474.
4. Codex Alimentarius Commission. Codex Standard 118-1979 (rev. 2008), Foods for
special dietary use for persons intolerant to gluten. Codex Alimentarius.
FAO/WHO, Rome, 2008.
RIDASCREEN® Total Gluten R7041
Niklas Weber, Lukas Kraft, Markus Lacorn, Thomas Weiss
R-Biopharm AG, Darmstadt, Germany
Introduction
The RIDASCREEN® Gliadin R7001 from R-Biopharm AG is based on the
monoclonal R5 antibody and has been endorsed as Codex Alimentarius Type I
method, AOAC Official MethodTM of Analysis 2012.01 Final Action and AACCI
approved method 38-50.01 [1-3]. The main epitope of the R5 antibody is the
pentapeptide QQPFP [4], which is present in many replicates in prolamins from wheat,
rye and barley; precisely α/β-, γ-, ω1,2- and ω5-gliadins from wheat, ω-, γ-40k- and γ-
75k secalins from rye as well as B-, C- and γ-hordeins from barley. The glutelins lowmolecular-
weight (LMW)-glutenin-subunits (GS) from wheat, high-molecular-weight
(HMW)-GS from wheat, HMW-secalins from rye and D-hordeins from barley are not
significantly detected by the R5 antibody.
Since the prolamins from rye and barley contain a higher copy number of the
pentapeptide QQPFP [5], the R5 antibody has a higher reactivity against rye and
barley compared with wheat, to which the RIDASCREEN® Gliadin R7001 is
calibrated to (PWG gliadin). Wheat is by far the most commonly used gluten
containing cereal in the world, so contamination of intended gluten-free products is
very likely to occur with wheat. The main exception to this is oats, which is usually
contaminated with barley, due to the geographic regions of cultivation, time of harvest
and further processing. This leads to frequent overestimations of the gluten content in
oat samples.
In order to address this issue, the AOAC has set up Standard Method Performance
Requirements (SMPR®) for the quantitation of wheat, rye and barley gluten in oats [6].
Due to the overestimation of the R5 antibody, the RIDASCREEN® Gliadin R7001
does not fulfil these requirements and the development of a new ELISA with a more
balanced quantitation of wheat, rye and barley was necessary.
Materials and methods
SMPR® 2017.021 reference materials [6] were obtained from Paul Wehling, General
Mills, Minneapolis, USA. These materials consist of a set of seven samples: (1) one
blank oat flour, (2) two oat flours spiked at levels of 10 mg/kg and 20 mg/kg wheat
gluten; (3) two oat flours spiked at levels of 10 mg/kg and 20 mg/kg rye gluten, and
(4) two oat flours spiked at levels of 10 mg/kg and 20 mg/kg barley gluten.
ELISA RIDASCREEN® Total Gluten (from R-Biopharm AG, Darmstadt, Germany)
was used according to instructions for use. This ELISA contains the R5 antibody, one monoclonal antibody raised against a known toxic sequence present on HMW-GS
from wheat and HMW-secalins from rye and two monoclonal antibodies raised against
a purified extract of LMW-GS proteins from wheat.
Purified gluten fractions LMW-GS, HMW-GS, rye prolamins and glutelins, barley
prolamins and glutelins were obtained from Katharina Scherf, Leibnitz Institute for
food system biology, Freising, Germany. The preparation of the material is described
elsewhere [7]. The material was solubilised in Cocktail (patented) (from R-Biopharm
AG, Darmstadt, Germany) and 80 % ethanol and diluted to suitable concentrations
according to the instructions for use of the RIDASCREEN® Total Gluten.
Results and discussion
For the development of the new ELISA, it was decided to keep the R5 antibody for its
high sensitivity to α/β-, γ- and ω1,2-gliadins from wheat, ω-, γ-40k- and γ-75k secalins
from rye as well as B-, C- and γ-hordeins from barley. Additionally, the R5 recognises
many peptides which were reported to be toxic for celiac disease patients [8, 9]. In
order to reduce the overestimation of rye and barley gluten in oats, additional
antibodies had to be combined with the R5 to counteract its high reactivity to rye and
barley.
Since another limitation of the R5 antibody is that it does not react with other relevant
gluten proteins (mainly glutelins), new antibodies against LMW-GS from wheat,
HMW-GS from wheat and HMW-secalins from rye as well as D-hordeins from barley
were raised. In the first attempt, reported toxic peptides from LMW-GS, HMW-GS
and from D-hordeins were selected and used for monoclonal antibody generation.
Different clones were obtained for all immunisations. However, only for HMW-GS, a
suitable clone was identified. For LMW-GS, a second immunisation yielded two
clones which in combination were suitable for LMW-GS detection. The clones for Dhordeins
turned out to be not sensitive enough for usage.
Characterisation of all antibodies included their reactivity against different gluten
fractions. For this, antibodies were used in homologue sandwich ELISAs (R5 as
capture antibody on the microtiter plate and as detection antibody in the conjugate,
LMW 1 on the plate and LMW 2 in the conjugate as well as HMW on the plate and in
the conjugate, respectively).
As expected, the highest reactivity of the LMW antibodies was against the LMW-GS
fraction from wheat (see Fig. 1A). Also the PWG gliadin showed an intermediate
reactivity to the antibodies. This is also not surprising, since the PWG gliadin contains
some wheat glutenins [10]. Additionally, the LMW-GS antibodies might also have a
weak cross-reactivity against wheat prolamins.
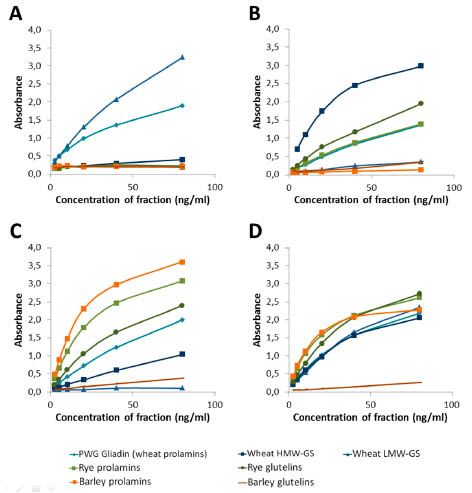
Figure 1. Reactivity of the (A) LMW 1 and 2 antibodies (B) HMW antibody (C) R5
antibody and (D) the combination of all four antibodies in the RIDASCREEN® Total
Gluten against different gluten fractions. Antibodies were used in separate sandwich
ELISAs for (A), (B) and (C), and combined for (D). Fractions from wheat are depicted
in blue colours (PWG in light blue, LMW-GS in medium blue and HMW-GS in dark
blue), rye in green (rye prolamins in light green and rye glutelins in dark green) and
barley in orange (barley prolamins in light orange, barley glutelins in dark orange).
The HMW antibody showed the highest reactivity against the HMW-GS from wheat,
followed by rye glutelins (see Fig. 1B). This was to be expected, as the immunisation
peptide for the antibodies contained a sequence present on HMW-GS from wheat and
HMW-secalins from rye, the latter being present in the rye glutelins fraction. The
minor reactivity against PWG gliadin was also expected, since the PWG gliadin contains some wheat glutenins [10]. The reactivity against the rye prolamins might
also be due to some contamination of this fraction by HMW-secalins. Additionally, the
HMW-GS antibody might also have a weak cross-reactivity against wheat and rye
prolamins.
The reactivity of the R5 antibody was as expected and previously reported [7] with
highest reactivity against prolamins from rye and barley (see fig 1C). Also rye
glutelins showed a high signal, probably due to γ-40k- and γ-75k secalins, which
contain the QQPFP sequence and are present in both prolamin and glutelin fraction
[personal communication by Katharina Scherf]. In general, the exact separation of
specific proteins into prolamins and glutelins in the course of the Osborne
fractionation is not completely possible, as co-precipitation and co-solubilisation
frequently occur [11, 12]. Intermediate reactivity was observed against PWG gliadin
and low reactivity against HMW-GS from wheat. The latter was not expected, but
might be due to some contamination by prolamin proteins.
In summary, each of the characterised antibodies recognised its target fraction with
highest reactivity, and since the LMW antibodies and the HMW antibody showed
highest reactivity against wheat fractions, these antibodies should be able to
compensate the overestimation of the R5 to rye and barley. With these four antibodies,
a combined sandwich ELISA was constructed with three antibodies combined in one
well (R5, LMW 1 and HMW) and three antibodies combined in one conjugate (R5,
LMW 2 and HMW). This combination was again tested for its reactivity against the
different gluten fractions (see Fig. 1D).
The combination of the antibodies showed a very well balanced reactivity against the
different gluten fractions. The only exception was the glutelins from barley (Dhordeins),
since no antibody was expected and able to detect this fraction. However,
the D-hordeins account for approx. 5 % of the barley gluten proteins only [12], so that
only a very minor component cannot be detected. The combination of the four
antibodies was further developed into a commercial product, the RIDASCREEN® Total Gluten with a 96 well microtiter plate coated with R5, LMW1 and HMW
antibody in each well, ready to use standards containing 0/5/10/20/40/80 mg/kg gluten
(standard material is an extract of four different wheat cultivars obtained from
Katharina Scherf), ready-to-use conjugate with R5, LMW 2 and HMW antibodies
conjugated to horse radish peroxidase, ready-to-use sample dilution buffer and a ten
times concentrated washing buffer. The overall incubation time is 50 min. The result is
given in mg/kg gluten as the sum result of prolamins and glutelins, thus the calculation
from prolamin to total gluten as in the RIDASCREEN® Gliadin is not necessary any
longer. This is a further advantage of this method, as the Codex factor of 2 for
calculation from prolamins to total gluten proteins is inaccurate in most cases and
leads to an overestimation [1, 13]. The extraction is performed using Cocktail
(patented) in combination with 80 % ethanol. Final dilution factor for samples is 1000.
The new ELISA was tested for its reactivity against the SMPR® reference material: oat
flours which were incurred with 10 and 20 mg/kg gluten from wheat, or rye, or barley, respectively [6]. As figure 2 shows, the new ELISA has a very balanced detection of
wheat, rye and barley. Three independent pilot lots of RIDASCREEN® Total Gluten
were produced and tested for their lot to lot comparison. All tested samples showed
very similar results in all three lots including the SMPR® reference material (data not
shown). Further in-house validation is ongoing. Preliminary results indicate a Limit of
Detection of approx. 2 mg/kg gluten and a Limit of Quantification of 5 mg/kg. More
than 80 potentially cross-reacting substances were tested, none was found to show
cross reactivity (data not shown).
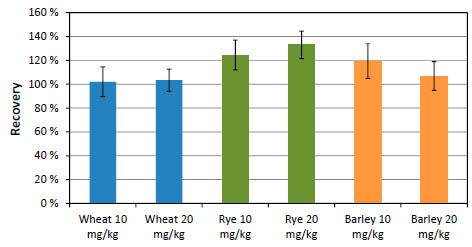
Figure 2. Reactivity of AOAC SMPR® samples [6] in RIDASCREEN® Total Gluten.
Each sample was extracted and analysed ten times. Wheat contaminated samples are
depicted in blue colour, rye in green and barley in orange.
An AOAC collaborative study with one of the pilot lots was performed in September
2018 with 19 laboratories worldwide using mainly oat samples. Preliminary results
showed very good lab to lab comparison (data not shown). After completion of the inhouse
validation, statistical analysis of the collaborative study and production of the
first lot in production scale, RIDASCREEN® Total Gluten will become commercially
available in 2019.
Conclusion
The new ELISA RIDASCREEN® Total Gluten combines the well-established
monoclonal R5 antibody with new antibodies against LMW-GS from wheat and
against HMW-GS from wheat and HMW-secalins from rye. It is thus the first
commercial ELISA targeting all major gluten fractions from wheat, rye and barley.
The detection of all relevant fractions prevents inaccurate quantitation due to the enrichment of a certain fraction during the processing of food, as it was reported for
e.g. starches [13-15]. The result is given in mg/kg gluten and is the sum result of
prolamins and glutelins; a calculation from prolamin content to total gluten content is
not necessary. The new ELISA shows a very well balanced detection of wheat, rye and
barley. The RIDASCREEN® Total Gluten is thus the ideal ELISA for analysis of oat
samples.
In addition to in-house validation, an AOAC collaborative study has been performed
in September 2018 using among other samples the SMPR® reference materials.
Preliminary results support the in-house validation data, in particular the balanced
detection of wheat, rye and barley.
Acknowledgement We would like to thank Nathalie Widmann for her tremendous amount of work in
developing this ELISA, the German Federal Ministry for Education and Research for
funding part of this work, Peter Köhler and Katharina Scherf for materials and
discussions, Paul Wehling for materials and discussions, Andreas Frey and Niels
Röckendorf for help with the characterisation of the antibodies and Maren Wiese for
testing materials.
References
1. 1. Codex Alimentarius Commission. Codex Standard 118-1979 (rev. 2008), Foods for
special dietary use for persons intolerant to gluten. Codex Alimentarius.
FAO/WHO, Rome, 2008.
2. Immer U and Haas-Lauterbach S, Gliadin as a measure of gluten in foods
containing wheat, rye, and barley-enzyme immunoassay method based on a
specific monoclonal antibody to the potentially celiac toxic amino acid prolamin
sequences: collaborative study. J AOAC Int 2012; 95(4), 1118-1124.
3. Koehler P, Schwalb T, Immer U, et al. AACCI approved methods technical
committee report: collaborative study on the immunochemical determination of
intact gluten using an R5 sandwich ELISA. Cereal Foods World 2013; 58(3), 36-
40.
4. Kahlenberg F, Sanchez D, Lachmann I, et al. Monoclonal antibody R5 for
detection of putatively coeliac-toxic gliadin peptides. Eur Food Res Technol 2006;
222(5-6), 78-82.
5. UniProt: https://www.uniprot.org/
6. Boison J, Allred L, Almy D, et al. Standard method performance requirements
(SMPRs®) 2017.021: Quantitation of wheat, rye, and barley gluten in oats. J AOAC
Int 2018; 101(4), 1238-1242.
7. Lexhaller B, Tompos C, Scherf KA, Comparative analysis of prolamin and glutelin
fractions from wheat, rye, and barley with five sandwich ELISA test kits. Anal
Bioanal Chem 2016; 408(22), 6093-6104
8. Tye-Din J, Stewart J, Dromey J, et al. Comprehensive, quantitative mapping of T
cell epitopes in gluten in celiac disease. Sci Transl Med 2013; 2(41), 41-51
9. Roeckendorf N, Meckelein B, Scherf K, et al. Identification of novel antibodyreactive
detection sites for comprehensive gluten monitoring. PLoS One 2017;
12(7), e0181566/1-e0181566/17.
10. Van Eckert R, Berghofer E, Ciclitira PJ, et al. Towards a new gliadin reference
material - isolation and characterization. J. Cereal Sci. 2006, 43(3), 331-341.
11.Wieser H, The precipitating factor in coeliac disease. Bailliere's Clin
Gastroent 1995; 9(2), 191-207.
12. Wieser H, Koehler P, Scherf K, Celiac disease and gluten. Elsevier Inc., London,
2014; p. 105 and 107.
13. Wieser H, Koehler P, Is the calculation of the gluten content by multiplying the
prolamin content by a factor of 2 valid? Eur Food Res Technol 2009, 229(1), 9-13.
14. Scherf KA, Wieser H, Koehler P. Improved quantitation of gluten in wheat starch
for celiac disease patients by gel-permeation high-performance liquid
chromatography with fluorescence detection (GP-HPLC-FLD). J Agric Food
Chem. 2016; 64(40), 7622-7631
15. Scherf KA, Impact of the preparation procedure on gliadin, glutenin and gluten
contents of wheat starches determined by RP-HPLC and ELISA. Eur Food Res
Technol 2016; 242(11), 1837-1848
Development of a gluten reference material suitable for
gluten analytical methods
Eszter Schall1, Lívia Hajas1, Kitti Török1, Zsuzsanna Bugyi1, Katharina Scherf2,
Stefano D’Amico3, Regine Schoenlechner3, Peter Koehler4, Roland Poms5, Sándor
Tömösközi1
1 Budapest University of Technology and Economics, Department of Applied
Biotechnology and Food Science, Research Group of Cereal Science and Food
Quality, Hungary
2 Leibniz-Institute for Food Systems Biology at the Technical University of Munich,
Freising, Germany
3 University of Natural Resources and Life Sciences, Department of Food Science
and Technology, Vienna, Austria
4 Biotask AG, Esslingen am Neckar, Germany
5 MoniQA Association, Güssing, Austria
Introduction
Coeliac disease is an autoimmune hypersensitivity reaction causing mucosal damage
and consequent absorption problems in the small intestine. The triggering components
are the gluten proteins found in some cereals (wheat, rye, barley). The only effective
treatment for the patients is a lifelong gluten-free diet [1]. Gluten-free (GF) products
are available for them with a regulatory threshold of 20 mg/kg gluten content [2].
Various analytical methods can be applied to measure gluten contamination in GF
products, but the most commonly used technique is the enzyme-linked immunosorbent
assay (ELISA). However, accurate gluten determination is hindered by several factors.
The protein content and composition of the cereals is not constant. The relative
proportions of gluten protein fractions vary depending on genetic (species, varieties)
and environmental factors (harvest year and agricultural practices). Additionally, their
physical (e.g. solubility, structure) and (bio)chemical (e.g. reactivity, affinity)
properties may change during food processing [3]. Commercially available ELISA
tests provide partly different strategies for determining gluten concentration as they
apply different extraction procedures, antibodies and target proteins and materials used
for calibration. The problem is that we have limited information about the effect of the
mentioned factors on the ELISA results and there is no certified reference material
(RM) to compare the different measurement results and to validate the gluten
analytical ELISA-based and alternative methods [4]. The production of a suitable RM
raises a few questions: Are different species, a single cultivar or a mixture of several
cultivars more suitable for this purpose? Is it necessary, and if yes, how can we take
into consideration the effect of environmental factors (i.e. the stability) of protein
composition? What form of protein sources - whole grain, flour or isolated protein - is
suitable for RM formulation? Our research group deals with the issue of gluten RM within the framework of international cooperation. Our aim is to investigate questions
related to the production of RM and to choose and produce a gluten RM candidate.
Materials and methods
23 different wheat cultivars were collected from all over the world to investigate the
effect of genetic variability. The applied workflow is demonstrated in Fig. 1. After full
protein characterisation of the samples, different quality and quantity criteria were
developed for selecting suitable varieties. These criteria related to the proper
quantitative range of crude protein content, gliadin/glutenin ratio, α-gliadin/γ-gliadin
ratio, gliadin recovery and the qualitative criteria related to the number of highmolecular-
weight glutenin subunits and separation of -gliadins. Five cultivars were
selected based on these criteria: Akteur from Germany, Carberry from Canada, Mv
Magvas from Hungary, Yitpi from Australia and Yumay-34 from China [5]. The
selected cultivars were collected from two harvest years (2014 and 2016) as well to
include environmental variability as an additional factor of the study. The five
different grains and their mixture of equal proportions were milled into white flours on
laboratory scale at the Budapest University of Technology and Economics, Hungary
(FQC 109 Micro-laboratory Mill, Metefém, Budapest, Hungary) and on pilot scale at
AGES, Austria (Bühler MLU-202 Laboratory Flour Mill, Switzerland). The gluten and
gliadin isolates were prepared from the flours on laboratory scale based on our
developed protocol: gluten isolates were produced with the use of the Glutomatic
System (Perten Instruments, Sweden), then gliadin was washed from dry gluten with
60 % (v/v) ethanol solution and freeze dried. Flours and protein isolates were
characterised by their protein content obtained by the Dumas method (N x 5.7), and
protein composition determined by SDS-PAGE, SE-HPLC [6] and RP-HPLC with
modified Osborne fractionation [7]. The gluten protein concentrations were quantitated
by RP-HPLC using PWG-gliadin as calibration reference [8]. The ELISA response of
the samples was determined using two commercially available ELISA test kits: the
AgraQuant Gluten G12 Assay (COKAL0200, Romer Labs, Tulln, Austria) and the
RIDASCREEN Gliadin Assay (R7001, R-Biopharm, Darmstadt, Germany).
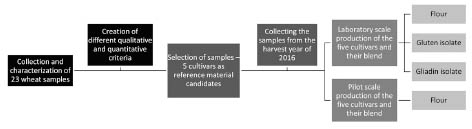
Figure 1. Flowchart for the experimental work on the development of lab and pilot
scale gluten reference material candidates
The analytical results were statistically evaluated with the investigation of means,
standard deviations, and factorial analysis of variance using Statistica 13 software
(StatSoft Inc., Tulsa, USA). Correlation analysis was carried out by linear Pearson
correlation at a confidence level of 0.95. The significance of differences was studied
using t-tests.
Results and discussion
The flours of the selected five cultivars from two harvest years show great variability
both in the crude protein contents and in the compositions according to separation
techniques. This variability also appears in the gliadin concentrations measured by
ELISA methods (Fig. 2). From the results, none of the varieties can be regarded as
completely average. The protein content of the flours was typically higher in the year
of 2016 than the year of 2014, so there were cultivars outside the quantitative
parameters of the selection criteria. This shows that the effect of harvest year has a
great influence on the amount of proteins and consequently the ELISA results, so it is
difficult to choose one cultivar with stable protein content. Similar gliadin content was
measured by the two ELISA methods, so using different methods has a smaller impact
than the effect of genetics or harvest year (Fig. 2).
The blended flour represents well the average of the five selected samples resulting in
a well-balanced genetic variability. Good homogeneity was obtained for it in each
examined parameter, indicating that the applied lab scale homogenisation method is
suitable for the production of blended flours. Additionally, the blended sample is best
suited according to the selection criteria.

Figure 2. Gliadin concentration of the five flour samples from two harvest years and
their blend from one harvest year measured by two different ELISA kits (methods A
and B)
An essential criterion for a RM is the availability of an adequate amount of product.
Therefore, upscaling of production was indispensable. According to our results, there
was no significant difference between the flours produced at laboratory and pilot scale
in the crude protein content and the ash content. Additionally, they had similar protein
composition and gliadin/gluten content measured by ELISA. These results proved that
we have succeeded in producing RM candidates in large scale (kg of material) which
are almost equal to the lab scale flours, not cross-contaminated and homogeneous
(data not shown).
As a second part of our work, gluten and gliadin isolates were prepared in laboratory
scale and compared to the flours of origin. The protein profiles of the isolates were
similar to the flours and there was no significant difference between the ELISA
recovery values of flours and their gluten isolates. Flours and gliadin isolates differed
significantly in just a few cases (Fig. 3). Therefore, loss of analytical information
should not be expected as a result of isolation.
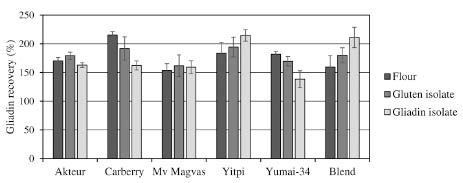
Figure 3. Gliadin recovery values of the six flours and their gluten and gliadin
isolates with ELISA kit A (calculation based on gliadin content measured by RPHPLC)
Conclusions
During our research we examined questions related to the production of a proper RM
for gluten analysis. After a preliminary selection process, five wheat cultivars were
chosen. Lab scale and pilot scale flours and lab scale gluten and gliadin isolates were
characterised by determination of protein composition and epitopes via ELISA
methods. Our results confirmed that the mixture of varieties can compensate the
genetic and environmental variabilities of individual varieties. Our upscaled
production procedure seemed to be successful, showing that we are able to produce
flour mixture-based reference candidate materials in pilot-scale.
In our experiments, flours and isolates gave similar results in protein composition and
ELISA response as well. Consequently, the decision about the application of flour over isolates in a RM should be based on other considerations (such as stability, solubility,
analytical application, etc.). Studies on these issues are still ongoing as a contribution
to solve this long-standing analytical problem of gluten RM.
Outlook
An international collaborative study will be organised to validate the selected flour
mixture and it is expected to be available for users in 2019. The last step of our
research in the production of wheat-based reference material is the production of a
processed incurred reference material modelling real food matrices and examining the
effects of food processing on the ELISA results.
Acknowledgement
The authors gratefully acknowledge the contribution of Prof. Ferenc Békés (FBFD
Pty. Ltd.), Dr. Terry Koerner (Head Allergen and Natural Toxin Section, Food
Research Division, Bureau of Chemical Safety, Health Canada) Dr. Marianna
Rakszegi (Agricultural Research Institute of the Hungarian Academy of Sciences) in
collecting the wheat samples and Dr. Elisabeth Reiter (Institute of Animal Nutrition
and Feeding, AGES, Vienna) in the pilot scale milling of the wheat samples. This
research is related to the scientific goals of the MoniQA Association. This work was
partially supported by the BME-Biotechnology FIKP grant of EMMI (BME FIKPBIO).
References
1. Scherf KA, Koehler P, Wieser H. Gluten and wheat sensitivities – an overview. J
Cereal Sci 2016; 67: 2-11.
2. Codex Alimentarius Commission. Codex Standard 118-1979 (rev. 2008), Foods for
special dietary use for persons intolerant to gluten. Codex Alimentarius.
FAO/WHO, Rome, 2008.
3. Hajas L, Scherf KA, Bugyi Z, et al. ELISA response and gliadin composition of
different wheat cultivars grown in multiple harvest years. Acta Alimentaria 2017;
46: 187-195.
4. Diaz-Amigo C, Popping B. Accuracy of ELISA detection methods for gluten and
reference materials: A realistic assessment. J Agric Food Chem 2013; 61: 5681-
5688.
5. Hajas L, Scherf KA, Török K, et al. Variation in protein composition among wheat
(Triticum aestivum L.) cultivars to identify cultivars suitable as reference material
for wheat gluten analysis. Food Chem 2018; 267: 387-394.
6. Gupta RB, Khan K, MacRitchie F. Biochemical basis of flour properties in bread
wheats. I. Effects of variation in the quantity and size distribution of polymeric
proteins. J Cereal Sci 1993; 18: 23-41.
7. Wieser H, Antes S, Seilmeier W. Quantitative determination of gluten protein types
in wheat flour by reversed-phase high-performance liquid chromatography. Cereal
Chem 1998; 75: 644-650.
8. Van Eckert R, Berghofer E, Ciclitira PJ, et al. Towards a new gliadin reference
material – isolation and characterisation. J Cereal Sci 2006; 43: 331-341.
Comparative reactivity of avenins from different pure
oat varieties to gluten R5 and G12 ELISA
immunomethods
Isabel Comino1, María Isabel Torres2, Ángela Ruiz-Carnicer1, Verónica Segura1,
Carlos Galera3, Carolina Sousa1, Ángel Cebolla3,4
1 Department of Microbiology and Parasitology, University of Seville, Seville, Spain
2 Departament of Experimental Biology, University of Jaen, Jaén, Spain
3 Hygiena Diagnostica, Seville, Spain
4 Biomedal S.L. Seville, Spain
Introduction
Gluten derived from wheat, barley and rye trigger autoimmune gut mucosal damage in
coeliac disease (CD). However, the role of gluten from oat (avenins) has been
controversial for many years, as evidenced by the lack of consensus in the legislation
of gluten-free products. The difference in type of oats used, their purity and study
designs have not allowed a clear answer as to whether oats are safe or not for all CD
patients [1]. To simplify the problem, it has been broadly accepted that pure oats are
safe for most CD patients, and contamination with other cereals is the main problem to
face [2,3]. However, it has been shown in different studies that some varieties of oat
may contain certain levels of peptides that can trigger immunoresponse in CD patients
or cross-reactivity in gluten immunomethods used for food labelling [1,4-7]. One of
the common assumptions by many stakeholders is that oats are naturally gluten-free
and only when oats are contaminated with other immunotoxic cereals as wheat or
barley, reactivity in gluten ELISA tests could be observed [2,3]. However, some pure
oats cultivars have shown significant cross-reactivity with G12, A1, Skerritt and R5
antibodies [1,4-6]. Some coeliac T-cell activating sequences from oat have been
identified [4,5,7] and some oat varieties have elicited early inflammatory events
typical of CD [3-5,7,8]. There are some clinical studies where gluten challenges were
made with oats that described adverse effects in some volunteers of the clinical studies
[2]. In this work, we showed that the cross-reactivity of some pure oat cultivars with
R5 or G12 antibodies significatively correlated with the immunogenicity estimated by
c0eliac T cell activation.
Materials and methods
Oats (Avena sativa L.) from cultivars designated OE717, OA729, OM719, OC723,
OH727, and OL715 (obtained from Spanish and Australian commercial sources) were
used in this work. The purity of the oat cultivars were checked by visual inspection
and PCR amplification with primers described elsewhere [1]. These cultivars were chosen based on their previously reported CD immunoactivity [1]. The immunoassays
were performed from the guidelines of the manufacturer (GlutenTox ELISA Sandwich
A1-G12 Hygiena Diagnostica, Seville, Spain; Ingenzim ELISA R5, Madrid, Spain).
Peripheral blood mononuclear cells (PBMCs) and cell cultures from 10 patients with
active CD on a gluten-containing diet were isolated from 6 ml of heparinised blood by
Histopaque gradient centrifugation and cultured at a density of 1X106 cells/ml in
RPMI-1640 culture medium. After 48 h, PBMCs were incubated with avenin, gliadin
and oryzenin peptides (50 g/ml).
Cell proliferation analysis and interferon gamma (IFN-) production were made
according to Comino et al. [1]. IFN- production by T cells with prolamin digests from
three different oat varieties. T lymphocytes were stimulated with digested prolamins
after treatment with tissue transglutaminase. IFN- production was evaluated by
ELISA after 48 h of incubation. The results are shown as the means of duplicate wells
and expressed as pg/ml. Gliadin and oryzenin were used as the positive and negative
control, respectively.
Results and discussion
We compared the reactivity of oat cultivars to the R5 and G12 antibodies (moAbs).
These seven oat cultivars, together with gliadin and oryzenin controls (Tab. 1) were
analysed for gluten content using sandwich R5 Ingezim and sandwich GlutenTox A1-
G12.
Table 1. Immunoreactive gluten content in different pure oat cultivars and control
samples estimated by ELISA R5 and G12 moAbs and by T cell immunogenicity
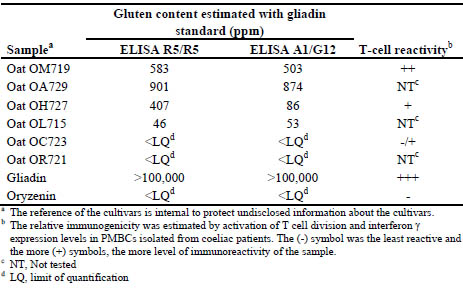
Conclusions
The immunogenicity of the analysed pure oat cultivars estimated by T-cell activation
correlated with the cross-reactivity to the main used monoclonal antibodies used to
detect gluten in food. Assuming the caution of the low number of analysed
immunogenic oats, the G12 reactivity correlated slightly more to the degree of
immunogenicity of the oat varieties because it could distinguish levels of differential
immunogenicity in the 3 out of 3 of the studied cultivars; 2 out of 3 in the case of R5.
The absence of reactivity to G12 antibodies may then indicate low risk of potential
immunotoxicity due to either the absence of toxic cereal contaminants or very low
content of immunogenic oat peptides.
References
1. Comino I, Real A, De Lorenzo L, et al. Diversity in oat Potential immunogenicity:
basis for the selection of oat varieties with no toxicity in disease. Gut 2011; 60:
915-922.
2. Fritz RD, Chen Y. Oat safety for celiac disease patients: theoretical analysis
correlates adverse symptoms in clinical studies to contaminated study oats. Nutr
Res 2018; 60: 54-67.
3. Hernando A, Mujico, JR, Mena MC, et al. Measurement of wheat gluten and
barley hordeins in contaminated oats from Europe, the United States and Canada
by sandwich R5 ELISA. Eur J Gastroent Hepatol 2008; 20: 545-555.
4. Real A., Comino I, de Lorenzo L, et al. Molecular and immunological
characterization of gluten proteins isolated from oat cultivars that differ in toxicity
for celiac disease. PLoS One 2012; 7: e48365.
5. Comino I, Bernardo D, Bancel E, et al. Identification and molecular
characterization of oat peptides implicated on coeliac immune response. Food Nutr
Res 2016; 60: 30324-30337.
6. Benoit L, Masiri J, del Blanco IA, Meshgi M, et al. Assessment of avenins from
different oat varieties using R5-based sandwich ELISA. J Agric Food Chem 2017;
65: 1467-1472.
7. Arentz-Hansen H, Fleckenstein B, Molberg Ø, et al. The Molecular basis for oat
intolerance in patients with celiac disease. PLoS Med 2004; 1(1): e1.
084-095.
8. Silano M, Penas-Pozo E, Uberti F, et al. Diversity of oat varieties in eliciting the
early inflammatory events in celiac disease. Eur J Nutr 2014; 53: 1177-1186.
Targeted LC-MS/MS reveals similar contents of α-
amylase/trypsin-inhibitors in all wheat species except
einkorn
Sabrina Geißlitz1, Christina Ludwig2, Katharina A. Scherf1, Peter Koehler3
1 Leibniz-Institute for Food Systems Biology at the Technical University of Munich,
Freising, Germany
2 Bavarian Center for Biomolecular Mass Spectrometry, Freising, Germany
3 Biotask AG, Esslingen am Neckar, Germany
Introduction
Among wheat hypersensitivities, non-celiac gluten sensitivity (NCGS) is least welldefined,
because the causative factors have not been unambiguously identified,
specific biomarkers are missing and the diagnosis relies on exclusion of coeliac
disease (CD), wheat allergies, other food intolerances and irritable bowel syndrome
[1]. The estimated prevalence ranges from 0.6 – 6 % of the population. The symptoms
occur several hours until a few days after consumption of wheat products and include
intestinal as well as extraintestinal (e.g., lack of wellbeing, tiredness, headache,
anxiety, foggy mind and joint/muscle pain) complaints. Upon treatment with a glutenfree
diet (GFD), the symptoms disappear [2]. In contrast to CD, NCGS patients have a
normal small intestinal mucosa, no autoantibodies and there does not appear to be a
relation to the expression of HLA-DQ2 or -DQ8, but they had increased numbers of
intraepithelial lymphocytes (IELs), levels of toll-like receptor (TLR) 2 and TLR4 and
reduced numbers of regulatory T cells, all markers of the innate immune response [3].
The pathomechanism of NCGS remains poorly understood. Potentially harmful wheat
components include gluten proteins, α-amylase/trypsin-inhibitors (ATIs), wheat germ
agglutinins and fermentable oligo-, di-, and monosaccharides and polyols
(FODMAPs). ATIs from gluten-containing cereals were found to induce the innate
immune response through activation of the TLR4-MD2-CD14 complex on monocytes,
macrophages and dendritic cells resulting in release of the pro-inflammatory cytokines
and chemokines IL-8, TNF-α and C-C motif chemokine ligand 2 (CCL2). ATIs were
also identified as adjuvants of pre-existing inflammatory adaptive immune responses
[4,5]. These ATI-induced innate immune responses are dose-dependent, suggesting
that a reduction of nutritional ATI intake may be sufficient to prevent inflammation.
Until the pathomechanism of NCGS is elucidated in more detail, NCGS patients are
advised to follow a GFD or a gluten-reduced diet.
ATIs make up about 2 - 4 % of wheat proteins and have been identified as major
causative factors of baker’s asthma. In the plant, they serve as defence proteins against
pests. Thirteen different ATI types (UniProtKB: 0.19, 0.28, 0.53, CM1, CM2, CM3, CM16, CM17, CMX1/3, CMX2, wheat subtilisin inhibitor, Bowman-Birk type trypsin
inhibitor and chymotrypsin inhibitor WCI) have evidence at protein level in common
wheat (Triticum aestivum L., hexaploid) [6]. Bioactivity assays with TLR4-expressing
monocytes showed that ATI extracts from spelt (T. spelta L., hexaploid), emmer (T.
dicoccum L., tetraploid) and einkorn (T. monococcum L., diploid) only had 30 – 70 %
of the activity of common wheat and it has been reported that einkorn had either very
low amounts of ATIs or even none [7,8].
All of the above led to the hypothesis that spelt, emmer and einkorn may be better
tolerated by NCGS patients, because of lower ATI contents compared to common and
durum wheat (T. durum L., tetraploid). To test this hypothesis, a targeted LC-MS/MS
method using stable isotope labelled peptides as internal standards (stable isotope
dilution assay, SIDA) was developed to quantitate the predominant ATIs 0.19, 0.28,
0.53, CM2, CM3 and CM16 in eight well-characterised common wheat, durum wheat,
spelt, emmer and einkorn cultivars, respectively.
Materials and methods
Grain samples
Eight cultivars each of common wheat, spelt, durum wheat, emmer and einkorn were
cultivated and harvested in 2013 by Friedrich Longin (State Plant Breeding Institute,
University of Hohenheim, Stuttgart, Germany) at Seligenstadt, Germany [9]. The
grains were milled into wholemeal flours using a cross-beater mill (Perten Instruments,
Hamburg, Germany) and analysed for water, ash, crude protein and albumin/globulin,
gliadin and glutenin contents [10].
Sample preparation for targeted LC-MS/MS and SIDA
Flour (50 mg) was extracted twice for 30 min at 22 °C with ammonium bicarbonate
(Abic) solution (0.5 mL, 50 mmol/L, pH 7.8). The suspensions were centrifuged for
25 min at 3750 × g, the supernatants combined and dried. The residue was dissolved in
Tris-HCl (320 μL, 0.5 mol/L, pH 8.5) and 1-propanol (320 μL). The five heavy
labelled peptides as internal standards (IS) were added (Tab. 1), followed by reduction
with tris(2-carboxyethyl)phosphine (TCEP), alkylation with chloroacetamide (CAA)
and lyophilisation. Tryptic hydrolysis (enzyme-to-substrate ratio 1:50) was performed
for 24 h at 37 °C in the dark. Having stopped the reaction with trifluoroacetic acid, the
solution was evaporated to dryness. The residue was dissolved in 0.1 % formic acid
(FA), filtered (0.45 μm) and used for targeted LC-MS/MS [11].
Targeted LC-MS/MS and SIDA
An UltiMate 3000 HPLC system (Dionex, Idstein, Germany) coupled to a triple-stage
quadrupole mass spectrometer (TSQ Vantage, ThermoFisher Scientific, Bremen,
Germany) was used.
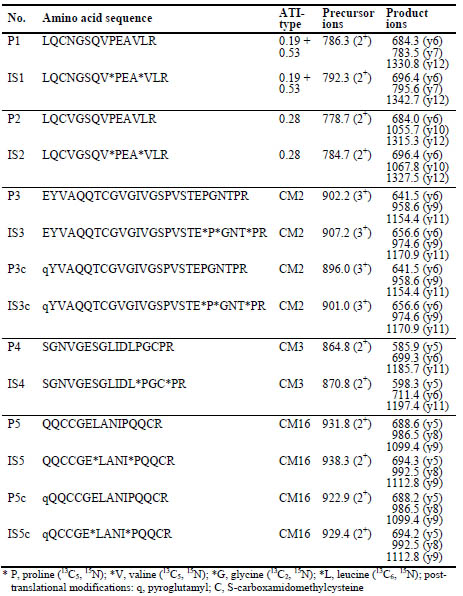
Table 1. Amino acid sequences of α-amylase/trypsin-inhibitor (ATI) marker peptides
(P) and internal standards (IS), and single reaction monitoring (SRM) parameters
(precursor ions and product ions).
Peptides were separated on an Aqua®-C18 column (50 × 2 mm, 5 μm, 12.5 nm,
Phenomenex, Aschaffenburg, Germany) with the following LC conditions: solvent A,
FA (0.1 %, v/v) in water, solvent B, FA (0.1 %, v/v) in acetonitrile; gradient, 0 - 5 min 10 % B, 5 - 20 min 10 - 90 % B, 20 - 23 min 90 % B, 23 - 25 min 90 - 10 % B, 25 -
40 min 10 % B; flow rate, 0.2 mL/min; injection volume, 10 μL; column temperature,
22 °C. The ion source was operated in the ESI positive mode with optimised source
parameters. Selected reaction monitoring (SRM) was used to analyse the transitions
from precursor to product ions (Tab. 1). Response lines were plotted by linear
regression of the peak area ratios A(P1-P5)/A(IS1-IS5) against the molar ratios
n(P1-P5)/n(IS1-IS5). Precision, limits of detection (LOD) and quantitation (LOQ) and
recovery of the method were determined as described by Geisslitz et al. [11].
Results and discussion
Method development for targeted LC-MS/MS and SIDA
First, untargeted LC-MS/MS analysis was used to identify the final five marker
peptides for ATIs 0.19+0.53, 0.28, CM2, CM3 and CM16 (Tab. 1). The peptides were
unique for each ATI type, except for P1 that occurred in 0.19 and 0.53. Suitable
precursor ions were identified by recording full-scan mass spectra of P1-P5 and
IS1-IS5 and the selected precursor ions were fragmented to define the three most
abundant transitions for selected reaction monitoring (SRM). The slopes of the
response lines with n(P)/n(IS) between 9.1 and 0.1 were between 0.7 and 1.2 and the
intercepts were close to 0.0, as expected for SIDA. Overall, the method showed good
performance characteristics in terms of repeatability (coefficient of variation, 2 - 5 %),
intermediate precision (coefficient of variation, 3 - 7 %), LODs (0.1 - 1.6 μg/g), LOQs
(0.3 - 4.7 μg/g), recovery assessed by spiking peptides into an analyte-free cassava
starch matrix (76 - 121 %) and recovery assessed by diluting common wheat flour with
cassava starch (92 - 102 %).
Application of the SIDA to the collection of flour samples (all ranges given as lowest
and highest values of the eight cultivars per wheat species) showed that the contents of
0.19+0.53 were significantly higher in common wheat (1.47 - 1.89 mg/g) and spelt
(1.51 - 2.08 mg/g) compared to durum wheat (0.84 - 1.15 mg/g) and emmer (0.90 -
1.25 mg/g). The contents of 0.28 were similar in common wheat (0.31 - 0.40 mg/g),
spelt (0.30 - 0.55 mg/g) and emmer (0.24 - 0.33 mg/g), but high variability was
observed in durum wheat, with two cultivars containing 0.21 mg/g (cv. LUN,
Lunadur) and 0.26 mg/g (cv. WIN, Wintergold) and the other six cultivars containing
amounts near the LOD of 6.6 μg/g. Emmer (0.37 - 0.58 mg/g) had the highest contents
of CM2, followed by durum wheat (0.29 - 0.54 mg/g), common wheat
(0.17 - 0.21 mg/g) and spelt (0.20 - 0.30 mg/g). The contents of CM3 were
significantly higher in spelt (1.07 - 1.59 mg/g), durum wheat (0.95 - 1.88 mg/g) and
emmer (1.32 - 1.98 mg/g) compared to common wheat (0.77 - 0.98 mg/g). Durum
wheat (1.15 - 2.07 mg/g) and emmer (1.38 - 2.10 mg/g) had higher contents of CM16
than common wheat (0.62 - 0.81 mg/g) and spelt (0.76 - 1.21 mg/g). This resulted in
overall ATI contents (sum of the five types) of 3.4 - 4.1 mg/g in common wheat
similar to durum wheat (3.2 - 5.5 mg/g), but lower than in spelt (4.1 - 5.7 mg/g) and
emmer (4.4 - 6.3 mg/g). The ATI contents of einkorn were between the LOD (40.5 μg/g) and 0.3 mg/g, thus confirming the absence or presence of very low
amounts of ATIs in einkorn.
The initial hypothesis was thus confirmed for einkorn, but refuted for spelt and emmer,
because the ATI contents were significantly higher than those of common wheat and
durum wheat. The contents of ATIs were not correlated to those of albumins/globulins
or crude protein. Principal component analysis (PCA) based on contents of each
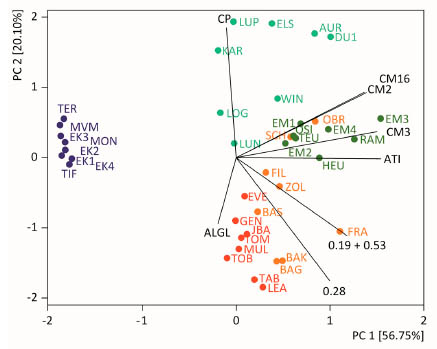
individual ATI, total ATI, albumins/globulins and crude protein showed that einkorn
cultivars distinctly clustered together opposite of the vector belonging to total ATI
contents (Fig. 1). Common wheat and spelt cultivars were mostly located in the lower
right rectangle, whereas emmer and durum wheat cultivars were located in the upper
right rectangle, with only two spelt cultivars in between. The ratio of CM-ATIs to the
sum of 0.19, 0.28 and 0.53 was about 1:1 in hexaploid wheat species, but around 3:1 in
tetraploid wheat species, so that the distribution of ATIs seemed to be suitable to
differentiate hexaploid from tetraploid wheat species.
Figure 1. Principal component analysis biplot of data for contents of single ATIs 0.19
+ 0.53, 0.28, CM2, CM3 and CM16, total ATIs (ATI), albumins/globulins (ALGL) and
crude protein (CP). Flours of eight common wheat (red), spelt (orange), durum wheat
(light green), emmer (dark green) and einkorn (dark purple) cultivars each grown at
the same location were analysed. PC, principal component. Figure modified from
[11].
Conclusions
The quantitative analysis of ATIs in five wheat species and eight cultivars each grown
under the same environmental conditions by targeted LC-MS/MS revealed that
einkorn contains very low amounts of the ATIs 0.19+0.53, 0.28, CM2, CM3 and
CM16. Compared to common wheat, spelt and emmer had higher contents of ATIs,
with durum wheat in between. Based on these results, it may be possible that einkorn
products are better tolerable for NCGS patients. Further in-depth investigations using
well-defined grain samples are necessary to substantiate this dataset and relate ATI
contents to in vivo bioactivity using markers of inflammation.
Acknowledgement
This research project was supported by the German Ministry of Economics and Energy
(via AiF) and the FEI (Forschungskreis der Ernährungsindustrie e.V., Bonn). Project
AiF 18355 N. The authors thank Sami Kaviani-Nejad, Ines Otte and Joerg Stein
(Leibniz-LSB@TUM) and Hermine Kienberger (Bavarian Center for Biomolecular
Mass Spectrometry) for excellent technical assistance. Special thanks go to Friedrich
Longin from the State Plant Breeding Institute (University of Hohenheim, Stuttgart)
for cultivating and providing the wheat samples used in this study.
References
1. Catassi C, Alaedini A, Bojarski C, et al. The overlapping area of non-celiac gluten
sensitivity (NCGS) and wheat-sensitive irritable bowel syndrome (IBS): an update.
Nutrients 2017, 9: 1268.
2. Molina-Infante J, Santolaria S, Sanders DS, Fernandez-Banares F. Systematic
review: noncoeliac gluten sensitivity. Aliment Pharmacol Ther 2015, 41: 807-820.
3. Fasano A, Sapone A, Zevallos V, Schuppan D. Nonceliac gluten sensitivity.
Gastroenterology 2015, 148: 1195-1204.
4. Junker Y, Zeissig S, Kim SJ, et al. Wheat amylase trypsin inhibitors drive
intestinal inflammation via activation of toll-like receptor 4. J Exp Med 2012, 209:
2395-2408.
5. Zevallos VF, Raker V, Tenzer S, et al. Nutritional wheat amylase-trypsin inhibitors
promote intestinal inflammation via activation of myeloid cells. Gastroenterology
2017, 152: 1100-1113.
6. Altenbach SB, Vensel WH, Dupont FM. The spectrum of low molecular weight α-
amylase/protease inhibitor genes expressed in the US bread wheat cultivar Butte
86. BMC Research Notes 2011, 4: 242-254.
7. Zoccatelli G, Sega M, Bolla M, et al. Expression of α-amylase inhibitors in diploid
Triticum species. Food Chem 2012, 135: 2643-2649.
8. Prandi B, Faccini A, Tedeschi T, et al. LC/MS analysis of proteolytic peptides in
wheat extracts for determining the content of the allergen amylase/trypsin inhibitor
CM3: Influence of growing area and variety. Food Chem 2013, 140: 141-146.
9. Longin CFH, Ziegler J, Schweiggert R, et al. Comparative study of hulled
(einkorn, emmer, and spelt) and naked wheats (durum and bread wheat):
agronomic performance and quality traits. Crop Sci 2015, 56: 302-311.
10. Geisslitz S, Scherf KA, Wieser H, Koehler P. Gluten protein composition and
aggregation properties as predictors for bread volume of common wheat, spelt,
durum wheat, emmer and einkorn. J Cereal Sci 2018, 83: 204-212.
11. Geisslitz S, Ludwig C, Scherf KA, Koehler P. Targeted LC-MS/MS reveals similar
contents of α-amylase/trypsin-inhibitors as putative triggers of nonceliac gluten
sensitivity in all wheat species except einkorn. J Agric Food Chem 2018, 66:
12395-12403.
Pathogenesis of coeliac disease: Identification of
isopeptides by LC-MS/MS
Barbara Lexhaller1, Christina Ludwig2, Peter Koehler3, Katharina Scherf1
1 Leibniz-Institute for Food Systems Biology at the Technical University of Munich,
Freising, Germany
2 Bavarian Center for Biomolecular Mass Spectrometry, Freising, Germany
3 Biotask, AG, Esslingen, Germany
Introduction
Coeliac disease (CD) is one of the most frequent food hypersensitivities affecting
approximately 1 % of the population worldwide [1]. This disease can be characterised
by three features: a) triggered by the ingestion of gluten, b) presence of the genetic
factor (HLA-DQ2 or DQ8), and c) generation of autoantibodies against tissue
transglutaminase (TG2) [2]. The gluten proteins are not sufficiently digested by the
human gastrointestinal enzymes, so long gluten peptides pass through the epithelial
layer and first trigger the innate immune response. Intraepithelial lymphocytes activate
defence mechanisms, which initiate apoptosis and increase epithelial permeability.
Secondly, TG2 modifies the gluten peptides by deamidation and transamidation. The
modified peptides stimulate gluten-specific T-lymphocytes, which finally lead to the
damage of the villi of the small intestine. Furthermore, antibodies are formed against
gluten peptides, TG2 and gluten peptide-TG2-complexes [2-4].
The two reactions of TG2 play a key role in the pathogenesis of CD. On the one hand,
the deamidation of specific glutamine residues to glutamic acid increases the immune
response. On the other hand, the transamidation and formation of gluten peptide-TG2-
complexes leads to the formation of antibodies against them. TG2 is a Ca2+-dependent
protein-glutamine γ-glutamyltransferase (EC 2.3.2.13), which catalyses the formation
of inter- and intramolecular Nε(γ-glutamyl)lysine bonds. The reactions take place in
the active site of the enzyme with the three amino acids cysteine-277, histidine-335,
and aspartic acid-358.
The aim of this study was to develop an analysis method by LC-MS/MS and to
identify isopeptides and their binding sites in the TG2-gluten peptide-complexes in a
model system.
Materials and methods
Model system of TG2 and the CD-active PepQ
The transamidation reaction of TG2 with the model peptide PFPQPQLPY-NH2 (PepQ)
was performed in 0.1 mol/l TRIS/HCl buffer (pH 7.4, 20 mmol/l CaCl2) at a molar
ratio of 1:150 at 37 °C for 120 min. To inactivate TG2, all samples were heated at 95 °C for 10 min. The samples containing the TG2-PepQ-complexes and the negative
controls were hydrolysed with trypsin and purified by solid phase extraction (SPE)
using 50 mg Sep-Pak tC18 cc cartridges (Waters, Eschborn, Germany). The
isopeptides and peptides were eluted with acetonitrile/water/formic acid (FA)
(40:60:0.1; 1 ml), dried and reconstituted in FA (0.1 %, v/v). For MS, the peptide
concentrations of the reconstituted samples were estimated with a NanoDrop Micro-
UV/VIS spectrophotometer (NanoDrop One, Thermo Scientific, Madison, USA) at
280 nm. The samples and negative controls were diluted in the 96 well plates to a
concentration of 200 ng/μL with acetonitrile/water/FA (2:98:0.1, v:v:v).
LC-MS/MS analysis was performed on an Ultimate 3000 nanoHPLC system (Dionex,
Idstein, Germany) coupled to a Q Exactive HF mass spectrometer (Thermo Fisher
Scientific, Dreieich, Germany). The nanoscale LC system is composed of a trap
column (75 μm x 2 cm, self-packed with Reprosil-Pur C18 ODS-3 5 μm resin, Dr.
Maisch, Ammerbuch, Germany) and an analytical column (75 μm x 40 cm, selfpacked
with Reprosil-Gold, C18, 3 μm resin, Dr. Maisch). The injection volume was
5 μL. The peptides were delivered to the trap column using solvent A0 (0.1 % FA in
water) at a flow rate of 5 μL/min and were then separated on the analytical column
using a 60 min linear gradient from 4 % to 32 % solvent B at a flow rate of
300 nL/min (solvent A1, 5 % DMSO, 0.1 % FA in water; solvent B, 5 % DMSO,
0.1 % FA in acetonitrile) [5]. The mass spectrometer was operated in data dependent
acquisition mode, automatically switching between MS and MS2 spectra. The mass-tocharge
(m/z) range of the acquisition of the MS1 spectra was 360 - 1300 m/z at an
Orbitrap full MS scan (60,000 resolution, 3e6 automatic gain control (AGC) target
value, 50 ms maximum injection time). In the MS2, peptide precursors were selected
for fragmentation by higher energy collision-induced dissociation (HCD; isolation
width of 1.7 Th, maximum injection time of 25 ms, AGC value of 1e5). Analysis was
performed using 25 % normalised collision energy (NCE) at a resolution of 15,000.
Data analysis of isopeptides
Data analysis was carried out with the Thermo Xcalibur .raw files in the MaxQuant
software (version 1.6.0.1) to search individual LC-MS/MS runs against the modelsystem-
specific database of α/β-gliadin. The TG2-isopeptide sites were configured as
modifications in MaxQuant by calculating the empirical formulas of the lysinecontaining
tryptic TG2-peptides (UniProt accession no. P21980). To use these peptides
as modifications (TG2-modifications) and to simulate an isopeptide bond formation, a
formal subtraction of NH3 was necessary. A theoretical protease for PepQ and the
model-specific database had to be configured with the following cleavage specificity:
QP, QY. For the individual search runs all parameters were used as default, except the
following settings: digestion mode: specific; maximum missed cleavage sites: 2;
variable modifications: every TG2-modification in one single search run; Fasta file:
UniProt accession no. P18573; fixed modifications: amidated C-term; minimum score
for modified peptides: 40; main search peptide tolerance: 4.5 ppm; mass tolerance for
fragment ions: 0.5 Da. The MaxQuant Viewer was used to confirm the identification of the isopeptides and the identification of the binding sites within the isopeptide by
assigning the b- and y-fragments of both sites to the same MS/MS spectrum.
Results and discussion
Identification of isopeptides
To identify the isopeptides in the model system, TG2 was incubated with PepQ. The
mix was then hydrolysed with trypsin, purified and subsequently measured by
nanoLC-MS/MS. The raw data were searched for PepQ with the TG2-modifications
and an identification score of > 100 was applied. With our data analysis strategy, we
were able to identify 22 different isopeptides. Fig. 1 presents the chromatogram of 21
isopeptides with the binding lysines indicated. For the isopeptide with the modification "K" it was not possible to illustrate the chromatogram for technical reasons.
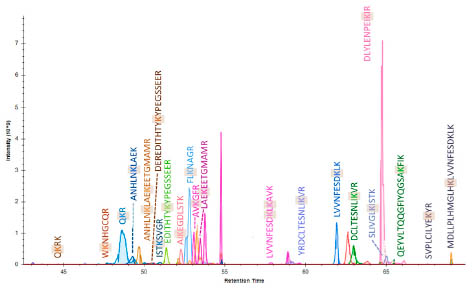
Figure 1. Chromatogram of 21 identified isopeptides. The TG2-site is shown,
respectively.
Within these 22 isopeptides, we were able to identify isopeptides with 6 known [6] and
11 new lysine binding sites of TG2. The 3D-model of the TG2 with its different
domains and the positions of the binding site is shown in Fig.2.
The three amino acids of the active site are marked in blue; they are located in the core
region. The lysines, which are involved in an isopeptide bond, are marked in orange
and are located more on exposed positions.
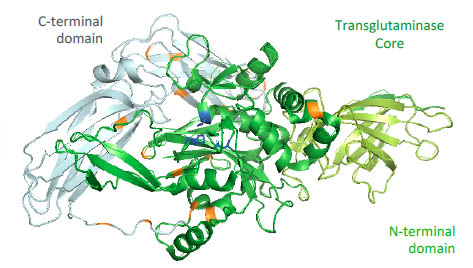
Figure 2. 3D-model of TG2 with the C-terminal domain, the core region and the Nterminal
domain. The amino acids of the active site are marked in blue, the identified
lysines are marked in orange.
The single isopeptides were identified with the data analysis strategy developed using
MaxQuant. The raw-files were searched from both sites, first against the TG2-
FASTA-file with PepQ-modifications and secondly against the PepQ-FASTA-file with
TG2-modifications. With the software tool MaxQuant Viewer it was possible to assign
the identified b- and y-fragments to the mass-spectrum.
For the annotation of both sides the same scan number was taken. In Fig. 3 two related
mass spectra are shown. On the upper left side the b- and y-fragments of the modified
PepQ are annotated to the spectrum and in the lower right corner the spectrum with the
fragments of the modified TG2-peptide LEAKEETGMAMR is displayed.
Conclusions
A software-supported and database-assisted proteomics search strategy with
MaxQuant and the search engine Andromeda to identify isopeptides between TG2 and
gluten-derived model peptides was established. Until now, six known [6] and 11 new
lysines of TG2 were identified as isopeptide binding sites.
Further work will focus on the improvement and advancement of the method for the
identification of isopeptides in gluten protein samples to identify the binding sites in
the gluten proteins of wheat, rye and barley.
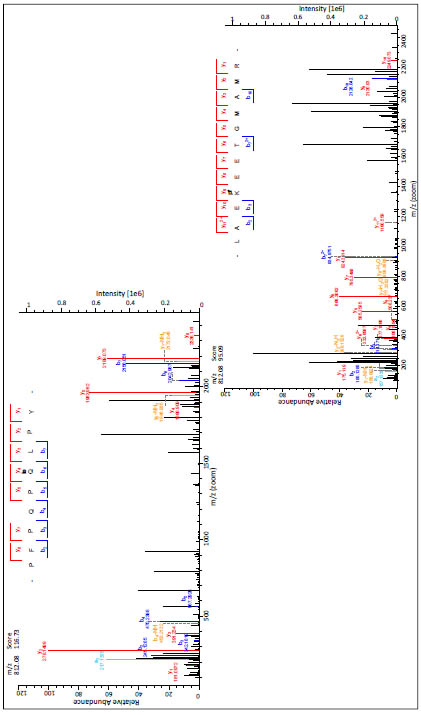
Figure 3. Mass spectra of the isopeptide (m/z = 812.08 (3+)) of the model peptide PepQ and the tryptic peptide LAEKEETGMAMR
of TG2
References
1. Ludvigsson J, Leffler, DA, Bai, J, et al. The Oslo definitions for coeliac disease
and related terms Gut 2013; 62: 43-52.
2. Schuppan D, Junker Y, Barisani D. Celiac disease: From pathogenesis to novel
therapies. Gastroenterology 2009; 137: 1912-1933.
3. Kagnoff M. Celiac disease: pathogenesis of a model immunogenetic disease J Clin
Invest 2007; 117: 41-49.
4. Qiao SW, Iversen R, Ráki M, et al. The adaptive immune response in celiac
disease. Semin Immunopathol 2012; 34: 523-540.
5. Hahne H, Pachl F, Riprecht B, et al. DMSO enhances electrospray response,
boosting sensitivity of proteomic experiments. Nat Methods 2013; 10: 989-992.
6. Fleckenstein B, Qiao SW, Larsen MR, et al. Molecular characterization of covalent
complexes between tissue transglutaminase and gliadin peptides. J Biol Chem
2004; 279: 17607-17616.
Wheat grain proteins with impact on end-use quality
and health attributes show significant responses on
heat, drought and combined stresses
Zsófia Birinyi1, Chris Florides2, Jäger Katalin3, Nándor Fodor4, Mariann Rakszegi5,
Ilma-Korponay Szabó6,7, Angéla Juhász1,2,
Gyöngyvér Gell1,8
1 Department of Applied Genomics, Agricultural Institute, MTA Centre for Agricultural
Research, Martonvásár, Hungary
2 Murdoch University, School of Veterinary and Life Sciences Perth WA, Australia
3 Plant Cell Biology Department, Agricultural Institute, MTA Centre for Agricultural
Research, Martonvásár, Hungary
4Crop Production Department, Agricultural Institute, MTA Centre for Agricultural
Research, Martonvásár, Hungary
5 Cereal Breeding Department, Agricultural Institute, MTA Centre for Agricultural
Research, Martonvásár, Hungary
6 Semmelweis University of Medicine, 1st Department of Pediatrics, Budapest,
Hungary
7 Coeliac Disease Center, Heim Pál Children’s Hospital, Budapest, and Department
of Pediatrics, Clinical Center, University of Debrecen, Debrecen, Hungary
8 Department of Applied Biotechnology and Food Science, Budapest University of
Technology and Economics, Budapest, Hungary
Abstract
In the view of global climate change, heat and drought stress have become the most
important limiting factors to crop productivity and food security. Beside this, there is
some information about the effect of the elevated atmospheric CO2 concentration to
the yield, but not on the quality. These abiotic factors, like water deficits and high
temperature generally occur at sensitive growth stages affecting functional properties
of wheat. They are reflected in changes in the wheat yield; adversely affect the seed
storage protein composition and the end-use quality. Wheat seed storage proteins are
considered as key players in triggering different wheat related health disorders,
containing immune responsive peptides (epitopes) that can cause immunological
diseases. The main aim of our research is understand the shifts in the seed storage
protein composition in different Hungarian bread wheat cultivars, caused by abiotic
stress factors using proteomics and immunomics analyses. Performing climate
chamber experiments and field trials we can evaluate the complex interpretation of
specific heat- drought and combined abiotic-stress effects and monitoring the influence
of elevated atmospheric CO2 on the composition of these proteins. Characterisation of
the protein content in the view of certain environmental conditions can provide a powerful tool to predict the effect of climate change on the bread making quality and
the accumulation of the harmful protein.
Introduction
In the past decades increased global temperature and climatic variability is resulting an
increase of both the frequency and magnitude of extreme climate events [1]. Climate
change can significantly affect environment, ecosystems, agriculture, and potentially
affect crop production and quality [2]. Therefore cereals might have exposed to more
than one stress event in the growing season [3].
With the rapidly changing climate combined abiotic stress factors can be detected, like
high temperature, lack of water in the root zone, extreme transpiration and elevated
atmospheric CO2, which can cause significant changes in the yield, relative protein
content, and composition of seed storage proteins, therefore affect the dough
rheological parameters, and baking quality [4,5,6].
As one of the most important crops providing vegetal protein in human food, wheat
plays a critical role in global food security. Storage proteins in wheat grain are the
single greatest source of protein in the human diet. Maintaining grain quality under
extreme climate conditions is crucial for human nutrition, end-use functional
properties [7].
Wheat seed storage proteins represent 70-80 % of the full protein content, depends on
species. They are key players in wheat-related diseases and health problems, like
autoimmune reaction of coeliac patients. They form a typical protein profile of modern
wheat genotypes, which confers to the dough its viscosity and elasticity. Bread quality
is determined by the composition and molecular structure of gluten which turn controls
the interaction of gluten subfractions during processing [8].
Abiotic stress factors and climatic extremities can cause changes in the protein profile
of seed storage proteins, due to shifts in the composition and elimination of their
balance.
The main purpose of the research was to observe the shifts and changes in the seed
storage protein composition of preselected Hungarian bread wheat cultivars, in
response to certain abiotic stress factors occurring at anthesis and post anthesis
periods.
Climate chamber experiments and field trials were performed to evaluate the complex
modelling of specific heat-, drought and combined abiotic stress effects and examine
the impact of elevated atmospheric CO2 on the wheat seed storage protein
composition.
Interpretation of the protein content in response to certain environmental conditions
can predict wheat acclimation, and effect of the climatic extremities on crop yield,
relative protein content, bread making quality and accumulation of the harmful
proteins in the future. Performing field trials with experimental free air CO2 enrichment (FACE) studies we
can build up an accurate climate prediction model, which can underlies a specific
breeding program.
Materials and Methods
Based on greenhouse heat and drought stress experiment out of 125 Hungarian bread
wheat genotypes, derived from the Cereal Gene Bank of the Department of Plant
Genetic Resources and Organic Breeding, Martonvásár, four wheat lines, a drought
resistant, a drought sensitive, a heat resistant and a heat sensitive were selected.
Grain yield, and plant height were measured, thousand kernel weight of all samples
were evaluated. Protein content was determined by near infrared spectroscopy (NIR)
and Duma method. Grain samples were milled with Retsch Mixer Mill MM200.
SE-HPLC was performed to analyse the profile of seed storage proteins by
determining the UPP% and Glutenin/Gliadin ratio. Harmful protein content was
determined by using commercially available R5 (RIDASCREEN® Gliadin, RBiopharm,
AG, Darmstadt, Germany) and G12 (AgraQuant Gluten G12 Assay (4-200
ppm) ELISA assays.
Results and Discussion
In reference to the previously selected four bread wheat genotypes, according to the
measured thousand kernel weight, combined drought and heat stress at anthesis and
post anthesis period caused a significant reduction, 40-50 % kernel weight loss, in
yield. However every genotypes shows increased protein content in response to
combined stress effects, based on the UPP% and glutenin/gliadin ratio, due to changes
in protein composition significant decrease can be found in quality of four of every
genotypes (Fig. 1).
The results of commercially available R5 and G12 ELISA tests revealed that based on
the climate chamber experiments characteristic changes occur in harmful protein
content in response to drought stress and heat stress at anthesis and post anthesis
period (data not shown). Further immunoanalyses are needed to evaluate different
abiotic stress profiles.
Besides the selected bread wheat genotypes Triticum monococcum preliminary
selected genotypes were examined by commercially available R5 and G12 ELISA
assays as well. They show significant lower harmful protein content compare to the
examined bread wheat genotypes.
Based on these findings further abiotic stress analysis and detailed examination of the
immunogen protein content will be needed to determine the stability of the lower toxic
protein content of the investigated einkorn genotypes.

Figure 1. Measured glutenin/gliadin ratio and UPP% (unextractable polymeric
glutenin proteins) of four Hungarian bread wheat and preselected Triticum
monococcum genotypes. C – control, DS A – drought stress at anthesis, HS A – heat
stress at anthesis, DS 6-10 DPA – drought stress at 6-10 DPA, HS 6-10 DPA – heat
stress at 6-10 DPA, COMB A – combined drought and heat stress at anthesis, COMB
6-10 DPA – combined drought and heat stress at 6-10 DPA
Conclusion
Due to the increasing impact of climate change, abiotic stress factors such as higher
temperature, lack of water, and elevated atmospheric CO2 can cause significant
changes in composition of seed storage proteins, therefore affect the dough rheological
parameters, and baking quality.
Performing climate chamber experiments and field trials complex interpretation of
specific heat, drought and combined abiotic stress effects can be evaluated, and the
influence of elevated atmospheric CO2 on the composition of these proteins can be
monitored.
Observation of the influence of abiotic stress factors on protein quality and
composition, and characterisation of the accumulation of harmful proteins provides us
a powerful tool to predict the effect of climate change on wheat end use properties, and
stability on response to certain environmental extremities. The results will lead us to contribute developing an accurate climate modelling program, which focuses on
complex factors of crop breeding.
Acknowledgement
This project was supported by OTKA PD 115641, GINOP-2.3.2-15-2016-00028,
János Bolyai Research Scholarship of the Hungarian Academy of Sciences, ÚNKP-18-
4-BME-393 National Excellence Program of the Ministry of Human Capacities and
MTA KEP-5/2016.
References
1. Yang F, Jørgensen A. D, Li H, Søndergaard I, Finnie C, Svensson B, Jiang D,
Wollenweber B, Jacobsen S, Implications of high-temperature events and water
deficits on protein profiles in wheat (Triticum aestivum L. cv. Vinjett) grain.
Proteomics 2011; 11(9): 1684-1695.
2. Change, I. C. The physical science basis. Contribution of working group I to the
fifth assessment report of the intergovernmental panel on climate change, 2013;
1535.
3. Dupont F. M, Altenbach S. B, Molecular and biochemical impacts of
environmental factors on wheat grain development and protein synthesis. J Cereal
Sci 2003; 38: 133-146.
4. Komatsu S, Kamal Ah. M, Hossain Z, Wheat proteomics: proteome modulation
and abiotic stress acclimation. Front Plant Sci 2014; 8(5): 684.
5. Fodor N, Diverging importance of drought stress for maize and winter wheat in
Europe. Nat Commun 2018; 9: 4249.
6. Högy P, Fangmeier A, Effects of elevated atmospheric CO2 on grain quality of
wheat. J Cereal Sci 2008; 48: 580-591.
7. Nuttall J. G, O’Learya G. J, Panozzoa J. F, Walker C. K, Barlowb K. M, Fitzgerald
G. J, Models of grain quality in wheat - A review. Field Crops Res, 2017; 202:
136-145.
6. Dhaka V, Khatkar B. S, Effects of gliadin/glutenin and HMW-GS/LMW-GS ratio
on dough rheological properties and bread-making potential of wheat varieties. J
Food Qual 2015; 38: 71-82.
5. Clinical research reports
High enzymatic digestibility of Triticum monococcum
gluten: evaluation of gluten stimulatory properties on T
lymphocytes from coeliac gut mucosa
Stefania Picascia1, Gianfranco Mamone2, Riccardo Troncone3,4, Carmen Gianfrani1,4
1 Institute of Protein Biochemistry, CNR, Naples, Italy
2 Institute of Food Sciences, CNR, Avellino, Italy
3 Department of Translational Medical Science, Section of Pediatrics, and
4 European Laboratory for the Food Induced Diseases, University of Naples
Federico II, Naples, Italy
Introduction
The ingestion of gluten proteins is the causative event triggering coeliac disease (CD)
in genetically predisposed individuals [1]. Because of the high content of prolin and
glutamine residues, gluten proteins are also termed as prolamines and represent the
main storage proteins in wheat, barley, and rye [2]. The peculiar chemical composition
confers to gluten proteins a high resistance to gastrointestinal digestion. As
consequences, large proline/glutamine rich peptides reach the small intestine and
trigger immune responses in patients with CD. Furthermore, the marked binding
affinity to CD-associated HLA molecules, consequence of a deamidation mediated by
tissue transglutaminase (tTGase), significantly increase the immunogenicity of gluten
[4]. Several peptides from α-, ω- and γ-gliadins have been described, to date, as
immunogenic in the great majority of both children and adults intolerant to gluten [5-
7]. The genetic susceptibility is conferred by HLA class II genes encoding for DQ2.5
or DQ8 heterodimers [8]. However, although 30 % of general population carry the
HLA DQ2.5/8 genes, it is estimated that only 1-3 % will develop CD in wheat
consuming countries [9]. The incidence of CD is much higher in the first-degree
relatives of coeliacs carrying the predisposing HLA-DQ genes, where the risk to
develop CD has been estimated 10-fold increased (approximately 10 %) [10]. Gluten
free diet is, currently, the only dietary therapy available for treatment of CD. However,
many alternative therapeutic strategies, based on different methodologies, are,
currently, under evaluation [11]. In the recent time an increasing interest has been
addressed, in particular, to the identification of wheat species naturally devoid, or with
a low content, of toxic gluten sequences [11-14], respectively suitable for treatment or
prevention of CD.
Triticum monococcum (TM), or Einkorn, is an ancient diploid wheat of special interest
to prevent CD. TM was the early wheat species to be cultivated by human beings, and
remained almost genetically unchanged since 10.000 years ago. Of note, TM gluten
proteins lack of a D-genome encoding the 33-mer, which is highly stimulatory of gluten-reactive T cells in coeliacs, as it contains several overlapping T cell epitopes
[13]. In addition, the good bakery and pasta-made, as well as nutritional, properties
make TM a good candidate for the treatment or prevention of CD [15]. However,
several studies, based on diverse experimental approaches, have showed conflicting
results on the immune-toxicity of TM gluten proteins [12-14, 16-19].
We have examined the capability of TM gluten to stimulate T cells from coeliac
patients after an extensive in vitro digestion, that reproduces the physiological gut
degradation [13,14]. The digested gluten peptides from TM and soft wheat Triticum
aestivum (TA), included as control, were used to in vitro challenge T-cell lines raised
from intestinal mucosa of CD patients.
Material and methods
In vitro simulated extensive gastrointestinal digestion
Gliadin samples from the wheats TM (Norberto-ID331 cultivar) and TA (Sagittario
cultivar) were enzymatically digested with sequential steps that included all proteases
from gastric-duodenal-brush-border membrane sections, as previously reported [20-
22]. Brush-border membrane enzymes (BBM) vesicles were isolated from pig jejunum
according to Shirazi-Beechey et al. [23]. Briefly, each gliadin preparation was
dissolved in 0.01 M chloride acid and incubated with pepsin (1:100, enzyme/substrate
w/w ratio) for 30 min at pH 2.0. The pH was next adjusted to 7.0 (by adding phosphate
buffer) and the gliadin mixtures were incubated with trypsin (1:100), chymotrypsin
(1:100), elastase (1:100), and carboxypeptidase A (1:500) for 1 hour at 37°C.
Thereafter, samples were two-fold diluted with 0.1 M sodium phosphate buffer pH 7.2,
supplemented with BBM at 100 mU/100 mg of peptide amount, and incubated at 37°C
for 6 hours (Fig. 1). Samples were stored at -80°C until further analysis.
Proteomic analysis of gastrointestinal resistant peptides and enzymatic deamidation
Gliadin proteins and peptides were identified using Nanoflow LC-ESI MS/MS
analysis as described [22]. Gastrointestinal resistant peptides, released upon the
extensive digestion, were fractionated by and HPLC and output peptide identifications
were validated by manual inspection of MS/MS spectra. The MS/MS raw spectra were
used to generate text files in mascot generic file format, and submitted to search
engines using Protein Prospector (http://prospector.ucsf.edu). Gliadin proteolytic
digests were next deamidated by tTGase (Sigma-Aldrich), as previously described
[20]. Gliadin samples were incubated at 37°C for 4-hrs with TG2 (1:1
enzyme:substrate ratio) in 5 mM Tris-HCl buffer (pH 6.8), containing 5 mM CaCl2,
10 mM NaCl, and 10 mM dithiothreitol. Samples were next lyophilised and store at -
20°C until the use on T cells.
Immunogenicity assay on intestinal T cells
T cell lines were established by repeated stimulations of intestinal cells isolated from
three HLA-DQ2.5 adult CD patients, with autologous mononuclear cells and PTgliadin
digest. Growing cells were thereafter expanded in culture with
phytohemagglutinin (PHA) and IL-2 and IL-15, as growth factors [12]. When in
resting phase, expanded T cell cultures were assayed for recognition of deamidated
gliadin enzymatic digests, as reported [12]. Briefly, 3x104 T cells were co-incubated
with 1x105 allogeneic and HLA-matched Epstain Barr virus-transformed B cells (BLCLs)
at 37°C in complete medium (X-Vivo enriched with 5 % human serum-Gibco,
Waltham, MA, USA) in 96-well plates. Gliadin enzymatic digests were used at 50 g/mL. Cell supernatants (50 μL) were collected after 48 hours of cell incubation for
the evaluation of INF-γ by a standard sandwich ELISA, as previously described [5].
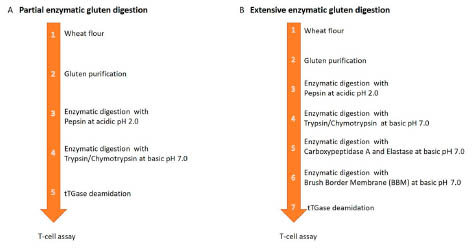
Figure 1. Schematic description of in vitro digestive process of gluten proteins and
functional assay on coeliac gut derived T cells. Digestion protocol widely used in most
scientific studies since 1980 that include a partial degradation with pepsin/trypsinchymotrypsin
enzymes (panel A). Extensive in vitro digestion comprehensive of all
gastro intestinal proteases, including the brush-border membrane enzymes (BBM)
(panel B).
Results and discussion
Gliadin extracts from ancient wheat species TM (Norberto-ID331 cultivar) and from
soft wheat TA (Sagittario cultivar) were in vitro enzymatically degraded, according to
an innovative protocol that aims to mimic the human physiological gastro-intestinal
digestion (Fig. 1). The proteomic analysis of digestion resistant peptides demonstrated
that gliadin proteins from the ancient wheat were more efficiently cleaved by GIenzymes
than gliadin proteins from the soft wheat (Fig. 2). As revealed by mass spectrometry analysis, a total of 76 peptides were identified as resistant to GI digestion
in TM gliadins, by contrast, 240 peptides were detected after the extensive degradation
of TA gliadin. Differences were also observed in the distribution of gastrointestinal
resistant peptides among the gliadin protein families. A high percentage of α-gliadin
(45 %) and γ-gliadin (46 %) peptides were found after the digestion of soft gliadin
(Fig. 2A), on the other hand, from the hydrolysis of ancient wheat the α-gliadin (66 %)
peptides were markedly prevalent (Fig. 2B). The sequence analysis of the
gastrointestinal resistant peptides revealed that the α-gliadin 33-mer and 31-55
peptides, respectively eliciting and T-cell mediated and innate immune response,
remained almost intact upon GI digestion of TA gliadin. Conversely, in the GIdigested
TM gliadin only the 31-55 peptides remained undegraded. Fragments of
DQ2.5-glia-α1a epitope (peptide 56-58 contained in 33-mer) and devoid of
immunogenicity, were released upon the GI proteolysis in both TA and TM gliadins
(Fig. 2C&D).
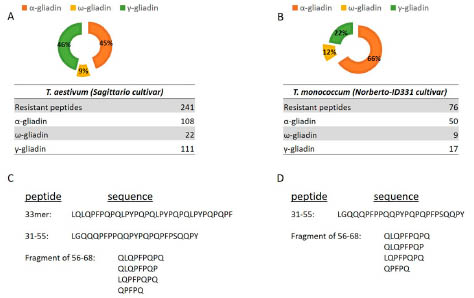
Figure 2. Profile of gliadin peptides resistant to the extensive in vitro digestion of
gliadin from soft wheat Triticum aestivum (Sagittario cultivar) (panel A&C) and from
ancient wheat Triticum monococcum (Norberto-ID331 cultivar) (panel B&D). Peptide
identification was obtained by the mass spectrometry analysis.
Overall, these data demonstrated that gliadin proteins from ancient and modern grains
have a dissimilar sensibility to GI degradation, that reflects the diverse genotype of
two wheat species and the different chemical structure of gliadins. In addition to the
proteomic characterisation, we next focused on the evaluation on how the extensive
digestion may affects the immunological properties of the two wheat species. Of note, previous studies reported conflicting results regarding the potential immune safety of
TM wheat for CD patients [12-19]. In order to have a clearer comprehension of the
immunogenicity of the ancient wheat, we evaluated the capability of TM gliadin to
activate coeliac gut T cells upon the physiologic GI proteolytic process. The detection
of the inflammatory cytokine INF-γ was used as sensitive probe to evaluate the
immune stimulatory potential of gliadins [5]. INF-γ production was assessed in T cell
lines previously obtained from the intestinal mucosa of three CD patients and highly
reactive to undigested TA gliadin [5,18,19]. In Fig. 3, the immune activation of gut T
cells in response to the GI-digested gliadin from both TA and TM wheats are shown.
Interestingly, we found that in all CD patients, the INF-γ production elicited by TM
gliadin was markedly reduced compared to the amount detected in response to TA
gliadin in all the TCLs assessed.
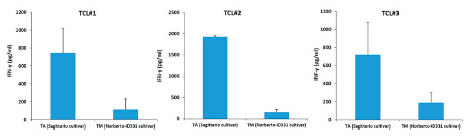
Figure 3. INF-γ responses obtained after in vitro stimulation of gliadin-reactive T cell lines
(TCL) established from intestinal mucosa of three coeliac patients. Gut T cells were in vitro
stimulated with the extensively digested gliadin from the soft wheat Triticum aestivum and
ancient wheat Triticum monococcum. Gliadin GI-digests were assayed at 50 μg/mL. The
INF-γ production was evaluated in cell supernatants after 48 hours of culture by enzymelinked
immunosorbent assay (ELISA).
These findings, obtained with coeliac gut T-cell assay, indicated that TM gliadin is
still toxic for coeliac patients, however after an in vitro extensive digestion retained a
reduced immunogenicity if compared to TA gliadin. The data on T cell reactivity
profile are in line with the proteomic analysis demonstrating a higher sensibility of TM
gliadin to be hydrolysed by gastrointestinal proteases that resulted in a reduced number
of immunogenic peptides.
Conclusions
In conclusion, we used an innovative in vitro protocol that mimic the physiological
gastrointestinal protein digestion, to evaluate the immunological properties of gluten
from the diploid wheat T. monococcum. Compared to gluten from the modern wheat,
such as the hexaploid T. aestivum, a low number of peptides remained undigested
upon the extensive digestion with gastro-intestinal proteases, including those of brushborder
membrane. The reduced amount of released peptides well correlated with the disappearance of the most immunogenic gliadin sequences and a low capability to
activate a T cells from coeliac gut mucosa.
Based on these findings, we suggested that the introduction of a wheat markedly
sensible to the GI digestion, might be of benefit for those patients at high risk to
develop CD, such as first-degree relatives of CD patients that carry the HLA genetic
predisposition. It would be of great interest to assess whether a prolonged T.
monococcum containing diet, may result in a reduction of CD incidence in at risk
subjects.
References
1. Stamnaes J, Sollid LM. Celiac disease: autoimmunity in response to food antigen.
Semin Immunol 2015; 27 (5): 343-352.
2. Koning F. Adverse effects of wheat gluten. Ann Nutr Metab 2015; 67 (2):8-14.
3. Lundin KE, Scott H, Hansen T, et al. Gliadin-specific, HLADQ(
a1*0501,b1*0201) restricted T cells isolated from the small intestinal mucosa
of celiac disease patients. J Exp Med 1993; 178 (1): 187-196.
4. Sjöström H, Lundin KE, Molberg O, et al. Identification of a gliadin T-cell epitope
in coeliac disease: general importance of gliadin deamidation for intestinal T-cell
recognition. Scand J Immunol 1998; 48 (2): 111-115.
5. Camarca A, Anderson RP, Mamone G, et al. Intestinal T cell responses to gluten
peptides are largely heterogeneous: implications for a peptide-based therapy in
celiac disease. J Immunol 2009; 182 (7): 4158-4166.
6. Tye-Din J, Stewart J, Dromey J, et al. Design of peptide-based immunotherapy and
diagnostics for celiac disease based upon comprehensive, quantitative mapping of
T-cell epitopes in gluten. Sci Transl Med 2010; 2 (41): 41ra51.
7. Hardy MY, Girardin A, Pizzey C, et al. Consistency in polyclonal T-cell responses
to gluten between children and adults with celiac disease. Gastroenterology 2015;
149 (6): 1541-1552.
8. Sollid LM. The roles of MHC class II genes and post-translational modification in
celiac disease. Immunogenetics 2017; 69 (8-9): 605-616.
9. Abadie V, Sollid LM, Barreiro LB, et al. Integration of genetic and immunological
insights into a model of celiac disease pathogenesis. Annu Rev Immunol 2011; 29:
493-525.
10. Gianfrani C, Troncone R and La Cava A. Autoimmunity and celiac disease. Mini
Rev Med Chem. 2008. 8 (2): 129-134.
11. Sollid LM, Khosla C. Novel therapies for coeliac disease. J Intern Med 2011; 269
(6): 604-613.
12. Suligoj T, Gregorini A, Colomba M, et al. Evaluation of the safety of ancient
strains of wheat in coeliac disease reveals heterogeneous small intestinal T cell
responses suggestive of coeliac toxicity. Clin Nutr 2013; 32(6): 1043-1049.
13. Molberg O, Uhlen AK, Jensen T, et al. Mapping of gluten T-cell epitopes in the
bread wheat ancestors: implications for celiac disease. Gastroenterology 2005;
128(2): 393-401.
14. Spaenij-Dekking L, Kooy-Winkelaar Y, van Veelen P, et al. Natural variation in
toxicity of wheat: potential for selection of nontoxic varieties for celiac disease
patients. Gastroenterology 2005; 129(3): 797-806.
15. Hidalgo A, Brandolini A. Nutritional properties of einkorn wheat (Triticum
monococcum L.). J Sci Food Agric 2014; 94(4): 601-612.
16. Vaccino P, Becker HA, Brandolini A, et al. A catalogue of Triticum monococcum
genes encoding toxic and immunogenic peptides for celiac disease patients. Mol
Genet Genomics 2009; 281(3): 289-300.
17. Zanini B, Petroboni B, Not T, et al. Search for atoxic cereals: a single blind, crossover
study on the safety of a single dose of Triticum monococcum, in patients with
celiac disease. BMC Gastroenterol 2013; 13: 92.
18. Gianfrani C, Camarca A, Mazzarella G, et al. Extensive in vitro gastrointestinal
digestion markedly reduces the immune-toxicity of Triticum monococcum wheat:
implication for celiac disease. Mol Nutr Food Res 2015. 59 (9): 1844-1854.
19. Gianfrani C, Maglio M, Rotondi Aufiero V, et al. Immunogenicity of monococcum
wheat in celiac patients. Am J Clin Nutr 2012; 96(6): 1339-1345.
20. Shan L, Molberg Ø, Parrot I, et al. Structural basis for gluten intolerance in celiac
sprue. Science 2002; 297: 2275-2279.
21. Iacomino G, Fierro O, D'Auria S, et al. Structural analysis and Caco-2 cell
permeability of the celiac-toxic A-gliadin peptide 31-55. J Agric Food Chem 2013;
61: 1088-1096.
22. Picariello G, Miralles B, Mamone G, et al. Role of intestinal brush border
peptidases in the simulated digestion of milk proteins. Mol Nutr Food Res 2015;
59(5): 948-956.
23. Shirazi-Beechey SP, Davies AG, Tebbutt K, et al. Preparation and properties of
brush-border membrane vesicles from human small intestine. Gastroenterology
1990; 98: 676-685.
Normalisation of serology in potential coeliac disease
subjects does not invariably lead to a full normalisation
of intestinal inflammatory signs
Valentina Discepolo1*, Mariantonia Maglio2*, Roberta Mandile2, Flavia Imperato2,
Luigi Greco2, Renata Auricchio2, Riccardo Troncone2
1 Department of Medicine, University of Chicago, Chicago (IL), USA
2 Department of Medical Translational Sciences & European Laboratory for the
Investigation of Food Induced Diseases, University Federico II, Naples, Italy
Introduction
Coeliac disease (CD) is an immune-mediated systemic disorder elicited by gluten and
related prolamins in genetically susceptible individuals, characterised by the presence
of a variable combination of clinical manifestations, CD-specific antibodies, HLADQ2
or HLA-DQ8 haplotypes, and enteropathy [1]. The severity of the intestinal
damage can range from villous atrophy (VA), typical of the overt disease, to minor or
absent histological alterations. The term potential coeliac disease (PCD) has been
coined to identify a subset of patients who present an increase in IgA anti-tissue
transglutaminase antibodies (anti-TG2) in the blood, but a normal intestinal
architecture [2]. Among PCD, some show a completely normal mucosa (Marsh0),
others display an increase in the numbers of intraepithelial lymphocytes (Marsh1). The
natural history of PCD has not been fully elucidated yet. Nevertheless, in our
longitudinal cohort, we observed that around one third of patients developed fullblown
disease with villous atrophy over a 9 years follow-up, while 67 % of them
showed a persistently normal duodenal architecture, despite the positive serology [3].
Among those who did not develop tissue damage, about one third showed persistently
elevated anti-TG2 IgA level, another third showed fluctuant levels of anti-TG2 with
transiently negative values, while the rest of them showed a persistent normalisation of
antibodies titers over the course of the follow-up [3]. Why only one third of PCD
patients develop full-blow disease remains to be assessed. Whether it is only a matter
of timing and all of them will eventually develop full-blown disease during the course
of their lives will require a longer and attentive follow-up. On the other hand, why
some individuals who displayed positive coeliac-specific serology, will then show a
full normalisation of the antibody titers and never evolve toward full-blown disease
remains unexplained. In this study we aimed at investigating the histological features
of this last category of PCD patients.
Patients and methods
In our cohort of over three-hundred PCD patients we identified 69 subjects who
became persistently seronegative during follow-up (69/330, 21 %). All of them
displayed positive anti-tissue tranglutaminase2 (anti-TG2) and/or anti-endomysium
(EMA) IgA antibodies in at least two different determinations and showed a normal
small intestinal architecture at the time of diagnosis. We considered as “seronegative” each subject persistently showing negative serum titers of anti-TG2 and EMA
antibodies for at least two years after the last positive detection. The detection of
positive titers of either one of these two antibodies at any time during follow-up was
considered an exclusion criterion. None of these subjects had a concomitant IgA
deficiency, thus there was no need to consider IgG class antibodies and/or antideamidated
gliadin (DGP) antibodies. Anti-TG2 antibodies and EMA were detected
by ELISA and immunofluorescence, respectively.
In each patient, esophagogastroduodenoscopy was performed and five biopsies were
taken from the duodenum, including one fragment from the bulb and four from the
distal duodenum. Moreover, 15 out of 69 seronegative PCD patients underwent a
second endoscopy of the upper gastro-intestinal tract, in most cases to investigate the
onset of clinical symptoms potentially related to CD. Thus, we collected a second
biopsy at time of normalisation of anti-TG2 IgA serum antibodies.
Four out of five fragments, including the one from the bulb, were fixed in 10 %
formalin, embedded in paraffin, and then stained with hematoxylin. The histological
and morphometrical analysis was performed by light microscopy. A villous height to
crypt depth ratio (V:H) above 2 was considered normal [4]. The Marsh scoring was
assessed upon counting the number of lymphocytes infiltrating the small intestinal
epithelium (IELs): the presence of less or more than 25 IELs per 100 enterocytes
defined a Marsh score of 0 or 1, respectively. All fragments were scored according to
Marsh criteria by an expert pathologist who was blinded to any serology results,
however the assigned score was based on the fragment with the worst
histopathological picture. One fragment for each patient was included in an optimal
cutting temperature (OCT) compound (Killik, Bio-Optica, Milan, Italy), stored in
liquid nitrogen and subsequently used for immunohistochemical staining using anti-
CD3+ and anti-TcR-γδ+ antibodies, as previously reported [5]. The number of stained
CD3+ and TcR-γδ+ IELs per millimetre of epithelium was determined. Cut-off values
of 34 CD3+ IELs and 3.4 TcR-γδ+ IELs/mm of epithelium, were used to discriminate
normal vs increased infiltration. In addition, intestinal anti-TG2 IgA antibody deposits
were evaluated by double immunofluorescence on five-micron OCT sections and
evaluated using a fluorescent microscope (Axioskop2- plus; Zeiss MicroImaging Inc,
Milan, Italy). The intensity of fluorescence was assessed based on the staining
intensity and distribution. Statistical analysis was performed using GraphPad Prism4
for Windows, version 4.03. Data were compared by Mann-Whitney or χ2 test. A pvalue
below 0.05 was considered to be statistically significant.
Results
In our cohort of 330 PCD patients followed-up over 10 years 69/330 (21 %) became
seronegative despite staying on a gluten containing diet (GCD). Most of them (59/69,
85 %) became seronegative within the first 24 months of follow-up. Each subject
defined as PCD received a small intestinal biopsy at time of first appearance of IgA
anti-TG2 antibodies in the serum that showed a normal mucosa (Marsh0) or slightly
infiltrated epithelium, without any sign of crypts hyperplasia and/or villous blunting
(Marsh1). Moreover for 15/69 seronegative PCD patients we had the chance to collect
a second biopsy at time of normalisation of serum anti-TG2 antibodies. Comparing the
histological features of the biopsies at time of diagnosis (t0) and upon normalisation of
coeliac-specific serology (t1), no significant change in Marsh scoring was found upon
anti-TG2 normalisation. In particular, Marsh score remained unaltered for 9/15 (60 %),
while it improved from M1 to M0 in 4/15 (26.7 %) subjects. Unexpectedly, it changed
from M0 to M1 in 2/15 (13.3 %) subjects.
Immunohistochemical analysis revealed a reduction in the number of TcR-γδ+ IELs,
but not of the number of CD3+ IELs upon normalisation of anti-TG2 antibodies serum
levels. A moderate, although not significant, decrease of the TcR-γδ/CD3 ratio was
also observed. Interestingly, even though most patients showed a decrease in the
numbers of CD3+ (60 %) and/or TcR-γδ+ (66.7 %) IELs, more than 30 % of them
retained signs of inflammation in the intestinal mucosa despite serology normalisation
and around 20 % of them showed even an increase of both TcR-γδ+ and CD3+ IELs.
Double immunofluorescence analysis performed to highlight the presence of intestinal
anti-TG2 antibodies’ deposits revealed that most patients maintained a local
production of CD-specific antibodies in the duodenal mucosa, despite the normalisation
of their serum levels. However, even though 54 % of subjects retained an
intestinal production of anti-TG2 antibodies, a significant lower rate of them (69.2 %
at t0 vs 30.8 % at t1, χ2 test p=0.05) showed a high intensity (>2) of such deposits.
Altogether, these data suggest that despite the disappearance of serum anti-TG2
antibodies, a significant proportion of PCD patients retained inflammatory signs in the
small intestinal mucosa, including CD3+ IELs (33 %), TcR-γδ+ IELs (55 %), TcR- γδ/CD3 (46 %) above the cut-off values and duodenal production of anti-TG2
antibodies (50 %).
Conclusions
This preliminary study provides evidence that a subset of PCD patients can develop a
persistent normalisation of anti-TG2 serum titres over time, despite being on a GCD.
Notably, even though the coeliac-specific serology normalises over time, most of these
patients continued displaying some signs of small intestinal inflammation or the
persistence of a local abnormal gluten-dependent mucosal immune response. More
specifically, independently from the level of infiltration of the small intestinal
epithelium at the time of diagnosis, most patients showed still abnormal numbers of TcR-γδ+ IELs upon normalisation of anti-TG2 antibodies, suggesting that some
alteration at the level of the epithelium might be still present and responsible for the
recruitment or the persistence of a pool of TcR-γδ+ IELs even in the absence of
peripheral signs of an ongoing gluten-dependent humoral response. What these factors
are remains to be assessed. Furthermore, the persistent production of intestinal anti-
TG2 antibodies in more than half of these seronegative PCD patients suggests that a
gluten-dependent immune response can still be ongoing locally at the level of the
duodenal mucosa. Notably, the magnitude of this response is significantly lower upon
normalisation of serology, suggesting that despite the persistency of an inflammatory
response in the duodenum, its magnitude decreases in parallel with the downregulation
of the humoral gluten-specific immune response. Whether these mucosal inflammatory
hallmarks of CD will persist on a GCD diet despite a prolonged antibody
normalisation in the serum, or whether they require longer time to disappear, remains
to be addressed. A longer follow-up will allow clarifying this aspect. Accordingly, we
suggest that patients who showed, during their lifetime, positive CD-associated
serology require a careful follow-up even upon seroconvertion (anti-TG2 and/or EMA
normalisation).
Together with a series of previously reported evidences, our results contribute to
support the idea that PCD is a heterogeneous population. It will be pivotal to identify
the different subsets of PCD as well as their clinical features. Furthermore, the
identification of biomarkers that can predict their natural history would be pivotal and
will help implementing the most appropriate management for these patients.
References
1. Husby S, Koletzko S, Korponay-Szabó IR, et al. European Society for Pediatric
Gastroenterology, Hepatology, and Nutrition Guidelines for the Diagnosis of
Coeliac Disease. J Pediatr Gastroenterol Nutr 2012; 54 (1): 136-160.
2. Ludvigsson JF, Leffler DA, Bai JC, et al. The Oslo definitions for coeliac disease
and related terms. Gut 2013; 62(1): 43-52.
3. Auricchio R, Tosco A, Piccolo E, et al. Potential celiac children: 9-year follow-up
on a gluten-containing diet. Am J Gastroenterol 2014; 109 (6): 913-921.
4. Kurppa K , Ashorn M , Iltanen S, et al. Celiac disease without villous atrophy in
children: a prospective study. J Pediatr 2010; 157: 373-380.
5. Paparo F, Petrone E, Tosco A, et al. Clinical, HLA, and small bowel
immunohistochemical features of children with positive serum antiendomysium
antibodies and architecturally normal small intestinal mucosa. Am J Gastroenterol
2005; 100 (10): 2294-2298.
Study of cell death in duodenal mucosa in active
coeliac disease
Carolina N. Ruera1, Federico Perez1, Luciana Guzman2, Lorena Menendez2, Laura
Garbi3, Fernando Chirdo1
1 Universidad Nacional de La Plata, Instituto de Estudios Inmunológicos y
Fisiopatológicos - IIFP (UNLP-CONICET), La Plata, Argentina
2 Servicio de Gastroenterología “Sor María Ludovica”, La Plata, Argentina
3 Servicio de Gastroenterología Hospital San Martín, La Plata, Argentina
Introduction
Coeliac disease (CD) is a multifactorial immune-mediated disease that develops in
genetically susceptible individuals after ingestion of a group of proteins derived from
wheat, barley and rye, commonly named gluten. CD is a chronic enteropathy
characterised by severe changes in the histology of proximal small intestine mucosa.
These changes include massive loss of enterocytes and villi, and crypt hyperplasia [1].
The increased enterocyte death has been explained by an uncontrolled apoptosis
induction due to excessive activation of intraepithelial lymphocytes (IELs) [2,3].
Apoptosis has been considered as the main pathway of enterocytes death, however this
mechanism has been only partially studied [4].
The Fas-Fas ligand (FasL) system has been considered the major pathways in inducing
enterocyte apoptosis [5]. FAS and FASL belong to the Tumor Necrosis Factor (TNF)
family. While FAS is expressed constitutively in different tissues it can also be
induced by proinflammatory cytokines, FASL is expressed mainly on activated T cells
and natural killer cells.
A different mechanism for enterocyte killing involved the recognition of stress signals
(MICA) [6] on enterocytes by activating receptor (NKG2D) presents in intraepithelial
cells has also been reported [7].
Pyroptosis is a lytic form of cell death distinguished from apoptosis produced as
consequence of the activation of the inflammasome platform. Particularly, caspases
1/11 cleave Gasdermin D (GSDMD) protein to generate a N-terminal domain, which
forms a pore after location on the plasma membrane [8]. Through these pores, small
molecules as some proinflammatory cytokines (IL-1) and intracellular contents, are
released [9,10]. Dysregulation of pyroptosis promoted by microbial infection and
danger signals is often associated with excessive inflammation leading to a number of
inflammatory diseases [11,12].
The aim of this work was to characterise the cell death pathways in intestinal mucosa
in CD enteropathy.
Materials and methods
Patients
Duodenal biopsies were collected from paediatric and adult patients during the routine
procedure for CD diagnosis in the Gastroenterology Units of Hospital Sor María
Ludovica (paediatric patients) and Hospital San Martín (adult patients).
CD diagnosis was based on histological findings in duodenal sections, serological
analysis and clinical signs. Individuals in the control group were subjected to
endoscopic study for other clinical conditions (mainly dyspepsia or diarrhoea). All of
them presented, at the time of evaluation, negative serology for CD markers and
normal duodenal histology. Ethic committees from Public Health Institutions approved
this work.
Experiments
Expression of caspase 1, 3, 8 and Gasdermin D (GSDMD) were assessed by Western
blot (WB) in protein extracts from whole biopsy. Proteins were separated by 12.5 % or
15 % polyacrylamide gels on SDS-PAGE. WB analysis was performed by incubating
with primary specific antibodies: β-Actin (Abcam, cat ab8227), Caspase 1 (Santa
Cruz, cat sc-56036), Caspase 3 (CellSignaling, cat 9662), Caspase 8 (CellSignaling,
cat 9746) and GSDMD (Santa Cruz, cat sc-376318). Secondary anti-mouse IgG-HRP
antibody conjugated (Abcam, cat ab6789) or anti-rabbit IgG-HRP conjugate (BioRad,
cat #1706515) were used. Visualisation of antibody reactivity was performed by
chemiluminescence, using a commercial reagent (GE Healthcare, cat RPN2232), and
they were revealed with Romek photographic reagents (Gamaxa SRL, Argentina)
according to the manufacturer's instructions. Image analysis for relative quantification
of optical density was performed with ImageJ software. The optical density in the band
of interest corresponding to the protein under study was quantified and normalised to
the value of β-actin.
TUNEL reaction and immunofluorescence microscopy (IFI) analysis were performed
on sections of paraffin-embedded tissues. For TUNEL reaction The Dead End
Fluorometric TUNEL system kit (Promega) was used, following the manufacturer's
instructions. Cell count on fluorescence images was performed using the ImageJ
software. The result was expressed as the number of TUNEL+ cells per μm2 of lamina
propria. Images were obtained using a Leica SP5 confocal microscope. The cell
density was calculated as the number of positive cells for each marker per unit area in
appropriate regions of tissue sections.
Statistics
Statistical analysis was performed using the GraphPad Prism 6.0 program. All
determinations between controls subjects and active CD patients were performed using
unpaired T-test with 95 % confidence, values of p <0.05 were considered significant.
Results and discussion
Using different markers and techniques, we aimed to characterise the cell death
process in the duodenal mucosa of untreated CD patients. First, we studied the
canonical apoptosis pathway, by analysing cell death by TUNEL reaction, caspase 8
and caspase 3 expression (Tab. 1).
Table 1. Evaluation of cell death pathways in duodenal mucosa
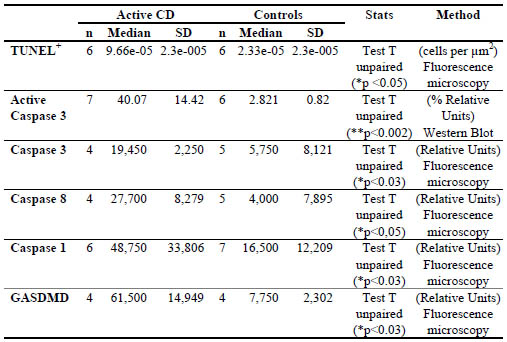
The TUNEL reaction, which represents an advanced stage in cell death, showed a
higher number of TUNEL+ cells of duodenal lamina propria in CD patients compared
to healthy controls. The massive number of lamina propria TUNEL+ cells scattered in
the tissue is intriguing. Identification of lineage of dead cells is in progress.
To evaluate the extrinsic pathway of apoptosis, caspase 3 and 8 were studied by
immunofluorescence analysis. Mean fluorescence intensity was higher for caspase 3
and 8 expression in duodenal samples of CD patients.
Since this evaluation cannot identify the active fraction of this protein, WB analysis
was performed to determine the cleaved caspase 3. WB analysis on total protein
extracts from whole duodenal biopsies showed increased levels of active caspase 3 in
samples from CD patients.
Finally, to assess whether pyroptosis may occur in the duodenum as part of the
pathogenic process, gasdermin D (GSDMD) was evaluated by immunofluorescence
assay on sections of duodenal samples. Levels of GSDMD were found statistically
higher in untreated CD patients compared with disease controls (Tab. 1).
Conclusions
In this study, we used immunofluorescence techniques and WB analysis to evaluate
critical components of cell death pathways in duodenal samples from untreated CD
patients and disease controls.
A high number of TUNEL+ cells, together with increased caspase 8 and caspase 3
levels in both epithelium and lamina propria cells were found in duodenal samples of
active CD patients. These findings are in agreement with previous descriptions about
apoptosis as a cell death pathway in the mucosa of untreated CD patients. In addition,
we showed activation of inflammasome/caspase 1/gasdermin D axis which suggest
that pyroptosis may also occur in this tissue.
In conclusion, we found that multiple pathways associated with cell death and
inflammation are activated in the small intestine of untreated CD patients. These
findings suggest that apoptosis, as well as pyroptosis, may occur jointly in the
enteropathy.
References
1. Sollid LM, Jabri B. Triggers and drivers of autoimmunity: lessons from coeliac
disease. Nat Rev Immunol 2013; 13(4): 294-302.
2. Ciccocioppo R, Di Sabatino A, Parroni R, et al. Cytolytic mechanisms of
intraepithelial lymphocytes in coeliac disease (CoD). Clin Exp Immunol 2000; 120:
235-240.
3. Pietz G, De R, Hedberg M, et al. Immunopathology of childhood celiac disease -
Key role of intestinal epithelial cells. PLoS ONE 2017; 12(9): e0185025.
4. Shalimar DM, Das P, Sreenivas V, et al. Mechanism of villous atrophy in celiac
disease: Role of apoptosis and epithelial regeneration. Arch Pathol Lab Med 2013;
137: 1262–1269.
5. Ciccocioppo R, Di Sabatino A, Parroni R, et al. Increased enterocyte apoptosis and
Fas-Fas ligand system in celiac disease. Am J Clin Pathol 2001; 115(4): 494-503.
6. Allegretti YL, Bondar C, Guzman L, et al. Broad MICA/B expression in the small
bowel mucosa: A link between cellular stress and celiac disease. PLoS ONE 2013;
8(9): e7365.
7. Hüe S, Mention JJ, Monteiro RC, et al. A direct role for NKG2D/MICA interaction
in villous atrophy during celiac disease. Immunity 2004; 21(3): 367-377.
8. Jorgensen I, Rayamajhi, M, Mioao EA. Programmed cell death as a defence against
infection. Nat Rev Immunol 2017; 17: 151-164.
9. Shi J, Gao W, Shao F. Pyroptosis: Gasdermin-mediated programmed necrotic cell
death. Trends Biochem Sci 2017; 42 (4): 245-254.
10. Ding J, Shao F. Growing a gasdermin pore in membranes of pyroptotic cells.
EMBO Journal 2018; 37: e100067.
11. Yuan YY, Xie KX, Wang SL, Yuan LW. Inflammatory caspase-related pyroptosis:
Mechanism, regulation and therapeutic potential for inflammatory bowel disease.
Gastroenterol Rep 2018; 6 (3): 167-176.
12. Masters, SL. Specific inflammasomes in complex diseases. Clin Immunol 2013;
147 (3): 223-228.
Deamidated gliadins worsen immune reaction in food
allergies
Clélia Villemin1*, Laure Castan1,2*, Marie Bodinier1*, Gregory Bouchaud1*
1 INRA, UR1268 BIA, Nantes, France
2 INSERM, UMR1087, Nantes, France
A food allergy, or hypersensitivity, is an abnormal response to an allergen that is
triggered by the immune system. The prevalence of food allergies varies from 4 to 8%
in children and from 1 to 5% in adults [1,2] and is constantly on the rise, making food
allergies a topical issue. Wheat is a major component of the human diet, particularly in
Western countries. Among food allergies, wheat allergy is predominant with an
estimated prevalence of 0.4% based on oral challenge experiments in American and
European populations [2,3]. Proteins derived from wheat grain have been implicated in
food allergies. In both children and adults, wheat allergy induces a variety of
symptoms: atopic eczema, atopic dermatitis, chronic urticaria, anaphylactic shock or
exercise induced anaphylaxis [4,5].
Wheat is composed of two fractions: the insoluble fraction and the soluble fraction.
The insoluble fraction is called gluten, and its proteins can be divided into gliadins and
glutenins. Wheat, particularly the gluten, contains a large variety of proteins that can
be allergens. Gluten has the particularity of forming a thick gel with great visco-elastic
properties. Gluten is largely used in the food industry, mostly because of its structure.
Indeed, it improves the texture and increases the protein quantity in food products.
However, its solubility restricts its utilisation as a food ingredient. To counteract this
problem, various processes such as deamidation (also called acid-hydrolysis), have
been used to improve its solubility. However, these processes can change the structure
of the proteins, and it has been shown that some processes produce new allergens [6].
Murine models constitute good tools to improve our understanding of allergic
sensitisation mechanisms and symptom elicitation [7,8]. The allergic response to
wheat proteins observed after a food allergy protocol is mostly orchestrated by Th2
lymphocytes. The sensitisation to some acid-hydrolysed wheat proteins increases the
production of IgE and IgG1 and the secretion of IL-4 and IL-5 by splenocytes [7]. In
the same way, sensitisation to deamidated gliadins induces the secretion of IgE, IgG1
and IL-4 [8].
Dendritic cells (DCs) are a key player in the establishment of oral tolerance and in the
induction of specific immune responses. In particular, specific populations of DCs in
the lamina propria are responsible for sampling soluble antigens in the intestinal lumen by extending their dendrites through the tight junctions of the intestinal epithelium and
transporting them to mesenteric lymph nodes (MLNs). This process is necessary for
the establishment of a specific immune response resulting in the activation of T and B
cells but also in the generation of regulatory T cells [9,10].
Even if the allergic response is well-documented, the exact mechanisms of the immune
response toward different forms of the same protein are still unclear. Indeed, how the
industrial processes modifying wheat proteins impact the immune mechanisms is
unknown. Our work aimed to study the effect of two forms of gliadins, native and
deamidated, on the sensitisation phase and the elicitation phase in a murine model of
food allergy. The immune response toward native gliadins and deamidated gliadins
was studied at three different time points: after the first sensitisation, after the second
sensitisation and after the oral challenge by gavage [11].
Materials and methods
Animal model
Balb/c mice from Janvier, France were used and housed in a ventilated cage system.
The protocol was approved by the Ethics Committee on Animal Experimentation of
Pays de Loire (CEEA) under the authorisation number 4049. Wild-type, 4-week-old
balb/cmice were sensitised to wheat native gliadins (NG) or deamidated gliadins (DG)
by two intraperitoneal injections of 10 μg in 100 μL of phosphate-buffered saline
(PBS) plus 100 μL of aluminium hydroxide and separated for 10 days. After one week,
mice were orally administered 20 mg of NG to induce the allergic reaction. Control
mice were sensitised intraperitoneally with aluminium hydroxide only and challenged
with PBS.
Gliadin preparation
Crude gliadins were extracted from wheat flour (Hardi cultivar) using the sequential
procedure. Briefly, the albumin–globulin fraction of the flour was removed by several
washings with saline buffer, and the flour was suspended in 70% ethanol for 1 h at
room temperature (RT) to solubilise the gliadins. After centrifugation (20 000 × g for
20 min at 41 °C), the supernatant, that is the whole NG extract, was collected and
freeze-dried. Deamidation was carried out by dispersing 20 mg of NG in 1 mL of 0.1
N hydrochloric acid. The temperature was increased to 90 °C and maintained at this
level for 1 h. The reaction was stopped by neutralizing with 0.1 N NaOH. The
resulting mixture was dialysed for 5 days against water (changed every day) and
centrifuged to recover the supernatant. This soluble material was then freeze-dried
(DG).
Measure of ear thickness
Just before the food allergy challenge, the ear thickness of the mice was measured
using a digital micrometre. One hour after gavage, the ear thickness was measured a second time. The results are expressed as the difference between the ear thickness
measured after the challenge and the ear thickness measured before it.
Immunoglobulin assay
Blood was removed by cardiac puncture 1 h after the challenge and centrifuged at
3000 rpm for 15 min. The quantification of gliadin-specific IgE was assayed on serum
samples by indirect ELISA as previously described [11,12,13].
Histology
One centimetre pieces of jejunum were extracted from mice, fixed in 4%
paraformaldehyde (PFA) for at least 48 h, embedded in paraffin and cut and stained
with hematoxylin eosin for morphological studies and inflammatory scoring.
Histological images were taken with an Axio Imager M1 microscope and an Axiocam
HRc (Zeiss). Sections were analysed using ImageScope for image acquisition (Aperio
Technologies, Inc.), and a cell type count for goblet cells and leukocytes was
performed with 10 villi per mouse. Grading of intestinal inflammation was determined
in a blinded fashion as previously described [14,15]. The total histological score
represents the sum of all features evaluated and thus ranges from 0 to 10.
Isolation of cells and flow cytometry
Mesenteric Lymph Nodes (MLNs) were collected from mice and ground. In all, 1.106
cells were transferred to a 96-well round-bottom plate and stimulated for 5 h with 50
ng/mL of phorbol-12-myristate-13-acetate and 1 μg/mL of ionomycin (Sigma-Aldrich)
together with brefeldin A (10 μg/mL; BD Bioscience). Cells were stained with surface
markers (CD3-FITC, CD4-Alexa 405, CD25-PE-Cy7, CD11c-BV510, MHCII-APCCy7,
SiglecH-PE, CD11b-APC, Ly6C-APCH7, CD103-BV4, CD68-BV4, Ly6G-PECy7,
B220-PerCP5.5, BD Biosciences) in the presence of Fc blocker mouse
CD16/CD32 antibody (BD Biosciences). For intracellular staining, the cells were fixed
and permeabilised using a Cytofix/Cytoperm kit (BD Biosciences) and stained with
IL-4-PE, Foxp3-APC, IFN-y-APC, RORγt-PE, or IL-17-APC-H7 (BD Biosciences).
Cells were analysed on a Canto II flow cytometer (BD Biosciences). Data were
acquired using Diva 8.0 software and analysed with FlowJo X (TreeStar).
Statistical analysis
Data were analysed using GraphPad Prism 6.0 (La Jolla, CA, USA). Values were
expressed as the mean ± SEM and compared using one-way ANOVA. In the case of a
significant ANOVA, means were compared using the Mann–Whitney U test, and the
results were corrected by Dunn’s multiple-comparisons test. A p-value of less than
0.05 was considered significant.
Results and discussion
To investigate whether development of allergic sensitisation and elicitation to different
wheat gliadin proteins is different, mice were sensitised and challenged with NG or
DG, using a validated protocol for food allergy (Fig. 1A) [14]. To characterise the
allergic symptoms, we measured the ear thickness with a micrometre after the first
sensitisation, after the second sensitisation, and after the oral challenge. The ear thickness
measured after each one of the sensitisations was the same for all the groups,
supporting the notion that it is the oral challenge that really triggers the allergic response
(data not shown). Our results show a significant increase in ear thickness after
the oral challenge for the mice sensitised with the deamidated form of gliadins compared
to mice sensitised with NG (101 vs 80 μm, p < 0.05) (Fig. 1B). The same results
were observed when measuring the level of wheat-specific IgE in the blood of mice
after the oral challenge (Fig. 1C). In fact, mice sensitised and challenged with deamidated
gliadins had higher levels of specific IgE compared to native gliadin allergic
mice (22 vs 14 mA.U, p < 0.001). At the tissue level, we assessed tissue inflammation
by hematoxylin-eosin staining of jejunum fractions (Fig. 1D). In line with the ear
thickness increase and specific IgE levels, mice allergic to deamidated gliadins exhibited
stronger alterations of the intestine compared to mice allergic to NG (4.13 vs 5.3,
p < 0.01) (Fig. 1E). Taken together, these results demonstrate that the sensitisation and
challenge with the deamidated form of gliadins strengthened the allergic response [11].
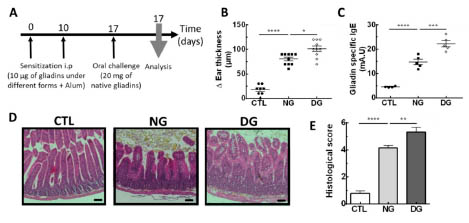
Figure 1. The deamidated forms of wheat gliadins amplify the allergic symptoms. A)
Mice were sensitised by intra-peritoneal injection of either DG or NG and then
challenged with NG by gavage. B) Measures of the ear thickness after the oral
challenge. C) Measures of wheat-specific IgE in serum of the three groups of mice
after the challenge. D) Hematoxylin-eosin stained jejunum sections from the CTL and
mice sensitised to NG or DG. E) Histological score calculated from jejunum sections
according to inflammation, crypt damage, and ulceration in three groups of mice.
Data represent the mean ± SEM (n = 8–10 animals per group); *p < 0.05, **p < 0.01,
and ***p < 0.001, ****p < 0.0001. Scale bar = 100 μm.
Our results are consistent with the observations in humans. In fact, the symptoms of
patients allergic to deamidated wheat proteins (anaphylactic shock, exercise-induced
anaphylaxis (ElA), urticaria) are often more severe than symptoms of patients allergic
to wheat (more often gut irritation, allergic rhinitis, asthma, and atopic
eczema/dermatitis syndrome (AEDS) than EIA and anaphylactic shock) [16,17]. Our
results demonstrate that deamidation induced by industrial processes provokes an
increase in allergic severity potential, as it has been suggested for peanut allergens
[18]. For peanuts, the allergic potential modification might be linked to protein
structure.
To characterise the immune response activated during sensitisation, we analysed cell
infiltration in the MLNs after the second sensitisation (Fig. 2). In line with our
previous results, the first sensitisation was not effective at inducing an immune
response as there was no increase in dendritic cell frequency in MLNs (data not
shown). In contrast, after the second sensitisation with DG, an increase of two- to
threefold was observed in the frequency of DCs in MLNs compared to native gliadinsensitised
mice (Fig. 2A). The NG group did not show an increase in dendritic cell
frequency compared to control mice. Transcriptomic, phenotypic, and functional
analyses indicate that human and murine DCs can be divided into three subsets: two
subsets of conventional DCs (cDC) called cDC1 and cDC2, and a third lineage of
plasmacytoid DCs (pDCs). Each subset is unique and has distinct functional specificities
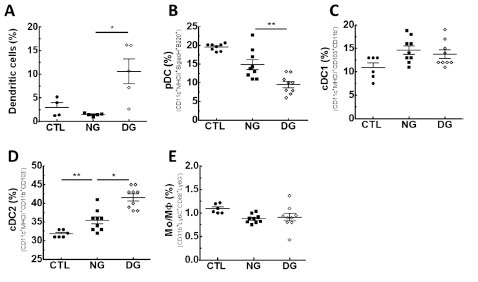
Figure 2. The deamidated form of gliadins induces influx of cDC and defect in pDC.
Frequency of A) DCs, B) pDC, C) cDC1, D) cDC2, and E) monocytes/macrophages in
the MLNs after the second sensitisation obtained by flow cytometry from the three
groups of mice. Data represent the mean ± SEM (n = 5–10 animals per group); *p< 0.05 and **p < 0.01.
to control T-cell responses. To examine which sub-population of DCs was modified,
we determined frequency of each population of DC. We showed that among DC,
pDCs were significantly decreased in DG sensitised mice compared to NG sensitised
mice (Fig. 2B). No difference was observed between NG-sensitised mice compared
with control mice (Fig. 2B). In contrast, frequency of cDC1 did not differ among the
different groups of mice (Fig. 2C). To the opposite, we observed an increase of cDC2
frequency in DG sensitised mice compared with NG sensitised mice (Fig. 2D). Finally,
the ratio of monocytes/macrophages frequency did not reveal any difference between
sensitised and control mice (Fig. 2E) [11]. To determine whether this increase in cDC2
DCs is associated with lymphocyte differentiation, we measured the Th1/Th2 and the
Th17/Treg ratio as they are known to be crucial factors in allergies (Fig. 3). As such,
after the first sensitisation, the ratio between CD3+CD4+IL-4+ Th2 cells and
CD3+CD4+IFNγ+ Th1 as well as CD3+CD4+IL-17+ Th17 cells and
CD3+CD4+CD25highFoxP3+ Treg cells were comparable among the four groups,
demonstrating that one sensitisation is not sufficient to induce immune response (data
not shown). In contrast, after the second sensitisation, the ratios of Th2/Th1 and
Th17/Treg were largely increased in mice sensitised with deamidated gliadins
compared to control mice. Moreover, the Th2/Th1 ratio remains unchanged in native
gliadin-sensitised mice compared to controls (Fig. 3A). To the opposite, Th17/Treg
ratio was significantly increased in DG sensitised mice compared with NG sensitised
mice (Fig. 3B). Overall, these results support that the deamidated form of gliadins
induces an earlier allergic immune response in mice than the native form of gliadins
[11].
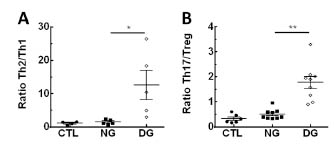
Figure 3. The deamidated form of gliadins induces Th2 polarisation and Th17
inflammation. A) Th1/Th2 and B) Th17/Treg ratios were measured in MLNs after the
second sensitisation and obtained by flow cytometry from the three groups of mice.
Th1 (CD3+CD4+IFNγ+), Th2 (CD3+CD4+IL-4+), Th17 (CD3+CD4+RORγt+IL-17+),
and Treg (CD3+CD4+CD25highFoxP3+) were measured after in vitro stimulation. Data
represent the mean ± SEM (n =5–10 animals per group); *p < 0.05 and **p < 0.01.
These results demonstrate the importance of pDC in tolerance establishment by
inducing Treg cells, as shown in [19]. Therefore, deamidation of gliadin induces a
modification in pDC influx and consequently a stronger sensitisation potential. This is
associated with a stronger and a faster presence of cDC2 known for its ability to prime naive T cells and to generate various effector T (Teff) cell phenotypes because of their
prominent expression of MHC class II and costimulatory molecules [20]. Results
demonstrate that the stronger sensitizing potential of deamidated gliadins is linked to
an earlier attraction of DCs (mainly cDC2) associated with a lack of pDC to
mesenteric lymph nodes which in turn induces an earlier Th2/Th17 response. Under
normal conditions, the Th1/Th2 cells are in a relatively balanced state in mice. When
body dysfunctions such as anaphylaxis occur, they are characterised by a shift in the
balance to Th2 cells, thereby resulting in a series of symptoms, including ear swelling,
increased gut permeability, and mast cell degranulation. Moreover, a decrease in Treg
cells and an inflammation linked to Th17 cells are also observed and participate to
inflammation and gut permeability in allergy. Our results were also consistent with the
observations of previous studies [21,22], verifying that industrial processing affects the
allergenicity of allergens in general.
Conclusions
This study focused on the potential for industrial processes to modify major wheat
allergens. The acid hydrolysis inducing deamidation produced greater allergenic
potential. In summary, our findings show that industrial processes could modify the
allergenic potential of gliadins by worsening its sensitizing capacity. Our data also
highlight that deamidation is sufficient for activating key immune pathways necessary
for sensitizing mice for immediate hypersensitivity reactions. The data demonstrated
the potential risk of partial hydrolysis of food proteins, in which hydrolysed proteins
may exhibit increased allergenic potency. Additionally, this experimental model using
Balb/c mice may be very useful for further basic and applied studies of the sensitizing
potential of various hydrolysed proteins.
References
1. Moneret-Vautrin DA. Allergic diseases. A very strong increase. Rev Prat.2007; 57:
1294-1296.
2. Nwaru BI, Hickstein L, Panesar SS, et al. Prevalence of common food allergies in
Europe: a systematic review and meta-analysis. Allergy 2014; 69: 992-1007.
3. Vierk KA, Koehler KM, Fein SB, et al. Prevalence of self-reported food allergy in
American adults and use of food labels. J Allergy Clin Immunol 2007; 119: 1504-
1510.
4. Battais F, Mothes T, Moneret-Vautrin DA, et al. Identification of IgE-binding
epitopes on gliadins for patients with food allergy to wheat. Allergy 2005; 60: 815-
821.
5. Jacquenet S, Morisset M, Battais F, et al. Interest of ImmunoCAP system to
recombinant omega-5 gliadin for the diagnosis of exercise-induced wheat allergy.
Int Arch Allergy Immunol 2009; 149: 74-80.
6. Denery-Papini S, Bodinier M, Larré C, et al. Allergy to deamidated gluten in
patients tolerant to wheat: specific epitopes linked to deamidation. Allergy 2012;
67: 1023-1032.
7. Adachi R, Nakamura R, Sakai S, et al. Sensitization to acid-hydrolyzed wheat
protein by transdermal administration to BALB/c mice, and comparison with
gluten. Allergy 2012; 67: 1392-1399.
8. Gourbeyre P, Denery-Papini S, Larre C, et al. Wheat gliadins modified by
deamidation are more efficient than native gliadins in inducing a Th2 response in
Balb/c mice experimentally sensitized to wheat allergens. Mol Nutr Food Res
2012; 56: 336-344.
9. Rescigno M, Urbano M, Valzasina B, et al. Dendritic cells express tight junction
proteins and penetrate gut epithelial monolayers to sample bacteria. Nat Immunol
2001; 2: 361-367.
10. Mazzini E, Massimiliano L, Penna G, et al. Oral tolerance can be established via
gap junction transfer of fed antigens from CX3CR1⁺ macrophages to CD103⁺ dendritic cells. Immunity 2014; 40: 248-61.
11. Castan L, Villemin C, Claude M, et al. Acid-Hydrolyzed Gliadins Worsen Food
Allergies through Early Sensitization. Mol Nutr Food Res 2018 ; 62: 1800159.
12. Bodinier M, Leroy M, Ah-Leung S, et al. Sensitization and elicitation of an allergic
reaction to wheat gliadins in mice. J Agric. Food Chem 2009; 57: 1219-1225.
13. Adel-Patient K, Creminon C, Bernardet H, et al. Evaluation of a high IgEresponder
mouse model of allergy to bovine beta-lactoglobulin (BLG):
development of sandwich immunoassays for total and allergen-specific IgE, IgG1
and IgG2a in BLG-sensitized mice. J Immunol Methods 2000; 235: 21-32.
14. Bouchaud G, Gourbeyre P, Bihouee T, et al. Consecutive Food and Respiratory
Allergies Amplify Systemic and Gut but Not Lung Outcomes in Mice. J Agric
Food Chem 2015; 63: 6475-6483.
15. Bihouee T, Bouchaud G, Chesne J, et al. Food allergy enhances allergic asthma in
mice. Respir Res 2014; 15: 142.
16. Leduc V, Moneret-Vautrin DA, Guerin L, et al. Anaphylaxis to wheat isolates:
immunochemical study of a case proved by means of double-blind, placebocontrolled
food challenge. Allergy Clin Immunol 2003; 111: 897-899.
17. Battais F, Richard C, Jacquenet S, et al. Wheat grain allergies: an update on wheat
allergens. Eur Ann Allergy Clin Immunol 2008; 40: 67-76.
18. Zhang W, Zhu Q, Zhang T, et al. Thermal processing effects on peanut allergen
Ara h 2 allergenicity in mice and its antigenic epitope structure. Food Chem 2016;
212: 657-662.
19. Uto T, Takagi H, Fukaya T, et al. Critical role of plasmacytoid dendritic cells in
induction of oral tolerance. J Allergy Clin Immunol 2018; 141: 2156-2167.
20. Uto T, Fukaya T, Takagi H, et al. Clec4A4 is a regulatory receptor for dendritic
cells that impairs inflammation and T-cell immunity. Nat Commun 2016; 7: 11273.
21. Beyer K, Morrow E, Li XM, et al. Effects of cooking methods on peanut
allergenicity. J Allergy Clin Immunol 2001; 107: 1077-10781.
22. Claude M, Lupi R, Bouchaud G, et al. The thermal aggregation of ovalbumin as
large particles decreases its allergenicity for egg allergic patients and in a murine
model. Food Chem 2016; 203: 136-144.
Kinetics and transcriptomics of gluten-specific T cells
after gluten challenge
Stephanie Zühlke1,3, Omri Snir2, Eivind Gard Lund1,3, Shiva Dahal-Koirala1,3, Ludvig
M. Sollid1,3, Knut E. A. Lundin1,4
1 KG Jebsen Coeliac Disease Research Centre, University of Oslo, Norway
2 Department of Clinical Medicine, University of Tromsø, Norway
3 Department of Immunology, University of Oslo, Norway
4 Department of Gastroenterology, Oslo University Hospital, Norway
Introduction
Coeliac disease (CD) is caused by an inappropriate immune response to gluten
proteins found mainly in wheat, rye and barley. Several types of T cells are involved in
CD: a) CD4+ T cells that recognise gluten epitopes presented by HLA-DQ2 or -DQ8
and which orchestrate responses of B cells and other T cells [1,2], and b)
intraepithelial lymphocytes (IEL) composed of CD8+ αβ T cells and γδ T cells that
induce tissue damage [1,3].
Concerning gluten specific CD4+ T cells, Anderson et al. have shown that after a
gluten challenge over 3 days in treated CD patients, interferon-γ producing, glutenspecific
cells measured by ELISPOT can be found on day 6 after challenge [4]. Gluten
specific cells CD4+ T cells can also be detected by HLA-DQ:gluten tetramers on day 6
after challenge [5-7]. Notably, HLA-DQ:gluten tetramer positive cells T cells express
gut-homing molecules like β7 integrin and CD103 [5] and the activation marker CD38
[8].
Despite several studies employing HLA-DQ:gluten tetramers [5-7], no kinetic analysis
of appearance of gluten-reactive T cells in blood after gluten challenge has been
performed. In addition, not much is known about functional aspects of HLADQ:
gluten tetramer staining T cells besides their cell surface markers which are
described above.
Since gluten-specific cells may in future be used for diagnostics and therapy outcome
[7,9], a more detailed understanding of their kinetics and cell biology is needed.
Hence, we aimed to study the kinetics and specific activation of gluten-specific CD4+
T cells.
Materials and methods
18 CD patients on a gluten-free diet underwent gluten challenge for 3 days with inhouse
developed, standardised gluten-containing cookies. Blood samples were drawn
prior to challenge. In the first study (kinetics study, n=8), cell kinetics in response to
gluten challenge were examined, and additional blood samples were drawn on day 1 and 4 – 8. For the second study (transcriptome study, n=10), study participants
donated blood samples on day 1 and 6 after the first intake of gluten. 6 participants
were challenged with only one cookie (1-day challenge) giving blood samples at
baseline and on day 6 after challenge (see figure 1).
Gluten-specific CD4+ T cells were stained with HLA-DQ:gluten tetramers and for the
kinetics study and 1-day challenge, their frequency was analysed on flow cytometer as
described elsewhere [7]. In the transcriptome study, gluten-specific CD4+ T cells
underwent subsequent cell sorting and RNA sequencing [9,10].

Figure 1. Timeline for the three gluten-challenge studies: the kinetics study and 1-day
challenge (above) and transcriptome study (below)
The studies are approved by the regional ethical committee of South-East Norway (ref.
2013/1237) and registered at clinicaltrials.gov (NCT NCT02464150).
Results and discussion
All participants gradually increased in tetramer+ cell numbers after gluten challenge
over 3 days, plateauing on day 6 to 8 (median 7 vs. 104 cells/106 CD4+ cells). 2
participants showed deviating kinetics. 1-day challenge was sufficient to increase
tetramer+ cells in all participants, yet resulting in a lower total cell number on day 6
(median 19 cells/106 CD4+ cells) (figure 2).
Flow cytometry and transcriptome analysis of gluten-specific gut-homing T cells
collected on day 6 after 3-day challenge revealed an upregulation of activation markers
(CD28, CD38, CD161, CD82), chemokine receptors (CCR9, CXCR3, CXCR6,
CCR6), gut homing markers (CD49, CD103) and IL-21.
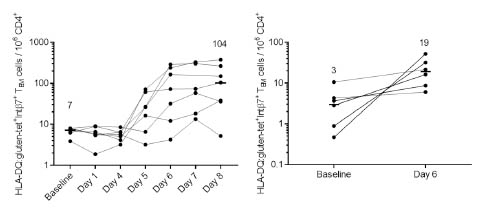
Figure 2. HLA-DQ:gluten-tet+ TEM cell kinetics (left) and cell numbers after 1-day
challenge (right)
Conclusions
This work provides insights into the kinetics and transcriptomic profile of antigenspecific
cells after gluten challenge. Sampling between day 6 and 8 after 3-day gluten
challenge is an appropriate time window for collection of gluten-specific T cells. Flow
cytometry and transcriptome analysis point to an activated, gut-homing phenotype of
gluten-specific T cells.
References
1. Sollid LM, Coeliac disease: dissecting a complex inflammatory disorder. Nat Rev
Immunol 2002; 2(9): 647-655.
2. Sollid LM, Iversen R, Steinsbø Ø, et al. Small Bowel, Celiac Disease and
Adaptive Immunity. Dig Dis 2015; 33: 115-121.
3. Jabri B, de Serre NP, Cellier C, et al. Selective expansion of intraepithelial
lymphocytes expressing the HLA-E-specific natural killer receptor CD94 in celiac
disease. Gastroenterol 2000; 118(5): 867-879.
4. Anderson RP, Degano P, Godkin AJ, et al. In vivo antigen challenge in celiac
disease identifies a single transglutaminase-modified peptide as the dominant Agliadin
T-cell epitope. Nat Med 2000; 6(3): 337-342.
5. Ráki M, Fallang LE, Brottveit M, et al. Tetramer visualization of gut-homing
gluten-specific T cells in the peripheral blood of celiac disease patients. PNAS
2007;104(8): 2831-2836.
6. Brottveit M, Ráki, M, Bergseng E, et al. Assessing Possible Celiac Disease by an
HLA-DQ2-gliadin Tetramer Test. Am J Gastroenterol 2011; 106(6): 1318-1324.
7. Sarna VK, Skodje GI, Reims HM, et al. HLA-DQ:gluten tetramer test in blood
gives better detection of coeliac patients than biopsy after 14-day gluten challenge.
Gut 2018; 67: 1606-1613.
8. du Pré MF, van Berkel LA, Ráki M, et al. CD62LnegCD38+ expression on
circulating CD4+ T cells identifies mucosally differentiated cells in protein fed
mice and in human celiac disease patients and controls. Am J Gastroenterol 2011;
106(6): 1147-1159.
9. Hardy M, Tye-Din, J. T cells in coeliac disease: a rational target for diagnosis and
therapy. Nat Rev Gastroenterol Hepatol 2018; 15: 583.584.
10. Picelli S, Faridani OR, Björklund ÅK, et al. Full-length RNA-seq from single cells
using Smart-seq2. Nat protoc 2014; 9: 171-181.
11. http://www.clontech.com/GQ/Products/cDNA_Synthesis_and_Library_Construction
/Next_Gen_Sequencing_Kits/Single_cell_RNA_Seq_Kits_for_mRNA_seq/Single_
Cell_RNA_Seq_v4.
Symposium Wheat Genomics
The first reference genome sequence for bread wheat
Angéla Juhász1,2*, Tatiana Belova3*, Iris Fischer4*, Daniel Lang4, Rudi Appels1, Odd-
Arne Olsen3, Klaus F. X. Mayer4, Manuel Spannagl4
1 State Agricultural Biotechnology Centre, School of Veterinary and Life Sciences,
Murdoch University, Murdoch, Western Australia, Australia
2 Applied Genomics Department, Agricultural Institute, Centre for Agricultural
Research, Hungarian Academy of Sciences, Martonvásár, Hungary
3 Norwegian University of Life Sciences, Aas, Norway
4 Plant Genome and Systems Biology, Helmholtz Zentrum München, Neuherberg,
Germany
Introduction
Wheat is one of the most important crops for human nutrition, but its components are
also causative for several human diseases and insensitivities, such as coeliac disease
(CD). With the recently published whole genome sequence for bread wheat [1], an
important resource is now available to identify and study gene families related to
wheat insensitivities and their products in a genome-wide manner. Its complex genetic
setup and large genome size of five times the human genome has made the wheat
genome impossible to decode for decades. The bread wheat genome consists of three
highly similar subgenomes (called the “A”, “B” and “D” subgenome, allo-hexaploid)
and has a repeat content of more than 80%, making it a grand challenge for short-read
sequencing and genome assembly. In 2005, the International Wheat Genome
Sequencing Consortium (IWGSC; https://www.wheatgenome.org/) teamed up with the
goal to produce a reference genome sequence for bread wheat by the year 2020.
Thanks to novel algorithms and bioinformatics strategies, a reference genome
sequence along with gene predictions could now be reported even earlier [1]. This new
resource established the foundation for in-depth analyses of gene expression and
regulation in the bread wheat genome [2] as well as the first genome-wide map of
genes and gene families encoding immunoresponsive proteins [3].
CD and wheat allergies such as Baker’s asthma and wheat-dependent exercise-induced
anaphylaxis (WDEIA) are the most common human diseases associated with wheat
grain products (as well as its relatives barley and rye). The majority of proteins
involved in these diseases belong to the group of proline-rich storage prolamins [4]
and are mostly encoded by gene families. CD is an inflammatory reaction to the
components of gluten (gliadins and glutenins) [5], whereas WEIDA and Baker’s
asthma are allergic reactions to either high-molecular weight glutenins (HMW glutenins) and omega-gliadins [6] or lipid transfer proteins (LTPs) and alpha-amylase
trypsin inhibitors (ATIs) [7], respectively. We therefore focused our research on those
gene families. This chapter will describe the progress in establishing high-quality and
comprehensive wheat genome resources and discuss the implications and possible
benefits for identifying genes and gene families involved in wheat insensitivities as
well as for selecting and improving wheat varieties with respect to their potential for
human health.
Materials and methods
Making use of the newly established wheat reference genome sequence, potential
allergens were identified and mapped in the wheat genome in a first step. For this,
members of the Pfam Prolamin clan (CL0482) – i.e. Gliadin (PF13016), LTP_2
(PF14368), Hydrophob_seed (PF14547), Tryp_alpha_amyl (PF00234), and
Prolamin_like (PF05617) – were used to identify gene members of the prolamin
superfamily. HMW glutenins containing the Glutenin_HMW (PF03157) Pfam domain
were also included. Finally, the pattern (-C-C-X n -C-X n -C-) was used to identify
small cysteine-rich sequences without a proper Pfam domain structure. The AllFam [8]
and ProPepper [9] databases were used to identify reference allergens and antigens and
to map annotated linear epitopes (i.e. immunoreactive protein sections) associated with
CD, WEIDA, and Baker’s asthma. Epitope toxicity was inferred from ELISPOT
assays used in a previous study [10]. Further details on the identification of potential
allergens and epitope mapping can be found in [3].
Second, a phylogenomic analysis was performed comparing bread wheat allergens to
nine other Poaceae species (Aegilops tauschii – the donor of the D genome, Triticum
urartu – the donor of the A genome, Secale cereale, Hordeum vulgare subsp. vulgare
cv. Morex, H. vulgare var. nudum, Brachypodium distachyon, Oryza sativa, Sorghum
bicolor, and Zea mays). Protein sequences were filtered and alignments and
phylogenetic trees were constructed as described in [1]. These protein sequences and
sequences from the UniProt database with PF13016, PF00234, and PF03157 domains
were used to further explore epitope prevalence and expansion in bread wheat and
related species. See [3] for more details.
Third, gene expression of allergens was evaluated in three different bread wheat lines
(Chinese Spring, Norwegian cultivars Bjarne and Berserk) under different temperature
conditions. Different grain tissues were extracted (transfer cells, aleurone cells, starchy
endosperm) and RNA isolation was performed as described in [11]. Details on
RNAseq, mapping, and trimming can be found in [3].
Results and discussion
The availability of a bread wheat reference genome sequence is a milestone in cereal
genomics. Its enormous size, high repeat content and hexaploid setup so far severely
complicated most genome-wide analyses. The lack of a reference genome sequence also had consequences for identifying genes and gene families underlying particular
traits or associated with certain characteristics such as baking quality. Thanks to novel
algorithms and bioinformatics strategies a true reference, chromosome-ordered
genome sequence has been reported for bread wheat recently [1], associated with a
high-quality gene prediction and annotation. This resource for the first time allows
comprehensive, reliable and genome-wide studies of individual genes and gene
families. In this context, a first genome-wide map of genes and gene families encoding
immunoresponsive proteins was generated [3] and published together with the
reference genome sequence.
To guarantee the quality of the annotation of genes encoding immunoresponsive
proteins, we performed extensive manual curation of gene models. In particular,
Prolamins contain recurring stretches of highly repetitive sequence, complicating
automated-only gene prediction severely. We identified 244, 321, and 229 genes
encoding immunoresponsive proteins on the A, B, and D subgenome, respectively. In
general, the immunoresponsive genes are located in conserved chromosomal areas,
usually in the telomeric region of the chromosome (Fig. 1). Thanks to a recent
common ancestry and high sequence conservation, this epitope map can also shed light
on immunoreative peptides in other cereal species such as barley and rye.
CD-associated epitopes with a high toxicity map mostly in the repetitive region of
alpha-, gamma-, omega-gliadins, and low molecular weight glutenins (LMW
glutenins) (Fig. 1). Peptides that show a high immunoreactivity are most abundant on
the D subgenome – indeed, the proteins showing the strongest immune response are
alpha- and omega-gliadins from this subgenome. Phylogenomic analysis revealed that
those highly toxic epitopes are only found in the A and D subgenomes, their donors (T.
uratu and Ae. tauschii), and rye. The major epitopes associated with Baker’s asthma
are ATIs located on chromosomes 3 and 6 of all subgenomes (Fig. 1). Those ATI
epitopes were found in all Triticeae. WEIDA-associated epitopes can be found in
HMW glutenins, gamma- and omega-gliadins – most notably on chromosome 1 in the
B subgenome. HMW glutenin-specific epitopes were found in all Triticeae, whereas
gamma-gliadin specific epitopes were only found in the bread wheat subgenomes and
the A and D donors. The results of the phylogenomic analysis show an emergence of
highly toxic epitopes very recently in the Poaceae evolution – and mostly in genes
involved in grain and baking quality.
The gene expression analysis showed that low temperature conditions reduced the
levels of toxic omega-gliadins. However, low temperature also increased the levels of
Baker’s asthma- and WEIDA-associated genes (mostly ATIs and LTPs). High
temperature, on the other hand, increased the expression levels of CD-associated genes
(mostly gliadins and glutenins). Additionally, expression levels differed significantly
between genotypes, especially for alpha-gliadins. Identifying effects of temperature
fluctuations on grain allergen content will be very helpful for further breeding
programs, especially with respect to the anticipated consequences of climate change.
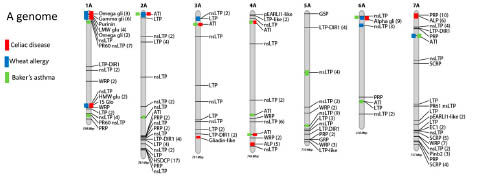
Figure 1. Distribution of coeliac disease (red), wheat allergy (blue), and Baker’s
asthma (green) associated genes over the seven A subgenome chromosomes of bread
wheat.
Conclusions
Wheat is one of the major food crops and has many positive nutritional aspects. A
small proportion of the human population, however, cannot safely consume wheat
grain products. The newly generated reference genome resources for bread wheat
allowed us to identify and map genes associated with CD and other wheat associated
diseases on the chromosome-scale genome sequence. This map is a first step to
enhancing breeding efforts of wheat lines that are saver for human consumption and
provide a higher environmental stability. Ongoing wheat pan-genomics and
resequencing efforts such as the 10+ wheat project
(http://www.10wheatgenomes.com) will greatly contribute towards understanding
natural variation and the possible impact of different wheat genotypes on the
immunoreactive potential of wheat products. Using a combination of genome
sequence data, epitope databases, in silico predictions, and gene expression data we
work towards a better understanding of the immunoreactive potential of wheat grain
end products.
Acknowledgements
The authors are grateful to the International Wheat Genome Sequencing and
Consortium (IWGSC) and Kellye Eversole for providing stimulating discussions in
developing this study.
References
1. International Wheat Genome Sequencing Consortium (IWGSC). Shifting the limits
in wheat research and breeding using a fully annotated reference genome. Science
2018; 361: eaar7191.
2. Ramirez-Gonzalez RH, Borrill P, Lang D, et al. The transcriptional landscape of
polyploid wheat. Science 2018; 361: eaar6089.
3. Juhasz A, Belova T, Florides CG, et al. Genome mapping of seed-borne allergens
and immunoresponsive proteins in wheat. 2018; Sci Adv 2018; 4: eaar8602.
4. Larre C, Lupi R, Gombaud G, et al. Assessment of allergenicity of diploid and
hexaploid wheat genotypes: Identification of allergens in the albumin/globulin
fraction. J Proteomics 2011; 74: 1279-1289.
5. Hardy MY, Tye-Din JA (2016) Coeliac disease: a unique model for investigating
broken tolerance in autoimmunity. Clin Transl Immunology 2016; 5: e112.
6. Scherf KA, Brockow K, Biedermann T, et al. Wheat-dependent exercise-induced
anaphylaxis. Clin Exp Allergy 2016; 46: 10-20.
7. Armentia D, de Luis J, Crespo L, et al. Wheat allergy. In: Watson RR, Preedy VR
(eds): Bioactive Food as Dietary Interventions for Arthritis and Related
Inflammatory Diseases. Elsevier Science, 2012, pp. 189-202.
8. Radauer C, Bublin M, Wagner S, Mari A, Breiteneder H (2008) Allergens are
distributed into few protein families and possess a restricted number of
biochemical functions. J Allergy Clin Immunol 2008; 121 (4): 847-852.e7
9. Juhász A, Haraszi R, Maulis C. ProPepper: a curated database for identification
and analysis of peptide and immune-responsive epitope composition of cereal grain
protein families. Database (Oxford) 2015; 2015: bav100.
10. Tye-Din JA, Stewart JA, Dromey JA, et al. Comprehensive, quantitative mapping
of T cell epitopes in gluten in celiac disease. Sci Transl Med 2010; 2: 41ra51.
11. Pfeifer M, Kugler KG, Sandve SR, et al. Genome interplay in the grain
transcriptome of hexaploid bread wheat. Science 2014; 345(6194): 1250091.
Gluten genomics in relation to editing coeliac disease
epitopes
M. J. M. (René) Smulders1, Aurélie Jouanin1, Luud J. W. J. Gilissen2
1 Plant Breeding, Wageningen University & Research, Wageningen, The Netherlands
2 Bioscience, Wageningen University & Research, Wageningen, The Netherlands
Introduction
Gluten are a complex mixture of various proteins in wheat, barley and rye. In wheat it
concerns glutenins and gliadins (prolamins). These are storage proteins located in
protein bodies in the grain endosperm, encoded by five gene families with up to
dozens of members each, clustered in loci on chromosomes 1 and 6. The loci include
genes with intact open reading frames, but also genes with premature stop codons,
which are considered to be pseudogenes [1]. Starting from one or a few genes in the
most recent common ancestor (MRCA) of all Aegilops and wheat species, their
number has expanded through tandem gene duplications and gene translocations [2].
The number of genes of the five gene families has now been established for the
hexaploid wheat variety Chinese Spring (with the AABBDD genome), of which the
whole genome has been sequenced recently using next generation sequencing
technologies [3].
Gluten forms a viscoelastic network that is essential for dough making, but gluten also
contains several immunogenic epitopes [4] that may cause coeliac disease in
genetically predisposed individuals. Coeliac patients have to follow a strict, life-long
gluten-free diet, which is difficult to adhere to, especially because wheat flour and
isolated (vital) wheat gluten is added to many processed food products for their
viscoelastic and binding properties. In addition, the nutritional value, the taste and the
overall quality of gluten-free products is often less than their wheat-based equivalents
(although these have considerable improved in quality in recent years, along with the
steady increase of the gluten-free market), and gluten-free products are more
expensive. This means that there is a need to develop coeliac-safe and healthy wheatbased
food products. Breeding wheat without the immunogenic epitopes would be a
definitive solution [5,6]. The recently developed CRISPR/Cas9 technology for
targeted mutagenesis can be a means to establish this.
Here, we review our application of CRISPR/Cas9 to modify gluten epitopes, or to
remove entire gluten genes or even complete gluten gene loci, in order to develop
coeliac-safe wheat. We start with recent results on the complexity of the gluten
genome and the way towards a high-throughput strategy in gluten gene identification
(GlutEnSeq) after modification. Further, if such modifications will become successful,
replacement of the current assays for total gluten detection in gluten-free products (to confirm a gluten content below the 20 ppm threshold) by an assay for the absence of
immunogenic epitopes (to confirm safe gluten) is needed. This will be discussed.
Finally, we will compare and discuss the results of gene editing approaches on gluten
genes by irradiation mutagenesis and by CRISPR/Cas technology from a European
legal perspective.
Gluten genomics
To apply CRISPR/Cas9 efficiently, detailed knowledge on the complete set of wheat
gluten genes is required. The genome of diploid Aegilops tauschii (the donor of the D
genome of bread wheat) has been sequenced and assembled [7-9]. Dong et al. [10]
studied a region of chromosome 1S (S: short arm) of Ae. tauschii, containing 103
genes among which several gamma-, delta- and omega-gliadin and LMW-glutenin
genes, with the corresponding (syntenic) region in Brachypodium and other
Gramineae genomes (rice, sorghum). The region in Ae. tauschii shared 17 prolamin
genes with Brachypodium but was 35 times larger compared to the syntenic region in
the Brachypodium, rice and sorghum genomes, including duplications of prolamin
genes and many insertions of retrotransposons. In hexaploid Chinese Spring the Ae.
tauschii-derived chromosome 1DS genome region was also much larger compared to
the corresponding regions on 1AS and 1BS [11]. The expansion of the genome size
during the evolution of the Triticum/Aegilops clade has not always led to the same type
of expansions, as the 6DS region of Chinese Spring, which includes alpha-gliadins,
was only half the size of the syntenic 6BS region [12].
The exact number of gluten genes in hexaploid wheat has long been unclear, with
estimations ranging up to 100-150 gene copies for alpha-gliadins alone, including
pseudogenes. The genome sequence of Chinese Spring [3] enabled a detailed study of
the number of prolamin genes present. Clavijo et al. [13] identified 105 full-length or
partial gluten genes (of which 29 alpha-gliadins, 18 gamma-gliadins and 10 omegagliadins,
6 HMW and 16 LMW) and 13 pseudogenes. This was based on an imperfect
annotation, even though it was manually curated, so the numbers may change.
Huo et al. [12] studied the Gli-2 loci, encoding alpha-gliadins, in Chinese Spring in
more detail, based on the genome sequence in combination with PacBio reads and
BioNano genome maps. They detected 10, 24, and 11 copies of the alpha-gliadins in
the A, B, and D genome, respectively. Among these, 26 encoded full-length proteins
and 21 (46%) were pseudogenes. Note that other hexaploid wheat varieties and
accessions will have different numbers of gluten genes as found in Chinese Spring.
Gluten gene editing with CRISPR/Cas
One can now use CRISPR/Cas9 to remove all gluten genes, which would produce a
gluten-free wheat which is interesting for many people who want to eat gluten-free,
but it would have an inferior baking quality. An alternative use of gene editing with
CRISPR/Cas9 is to precisely modify gliadin genes and strip them of immunogenic epitopes, to develop wheat with safe gluten. A couple of groups world-wide are
applying this technology to wheat to make it coeliac-safe. Sánchez-León et al. [14] in
Cordoba, Spain, successfully deleted complete gliadin genes in this way. Alternatively,
gliadin genes can be modified to precisely inactivate (by point mutation) or to delete
their immunogenic epitopes, which is the strategy in Wageningen, the Netherlands
(Jouanin et al., submitted). Next to these European groups, Calyxt (Roseville,
Minnesota, US) is working on gene-editing of wheat as well. Ultimately, commercial
varieties may be produced based upon a combination of these approaches, with some
gluten loci completely deleted and some loci present that only contain coeliac-safe
gluten genes, i.e., without epitopes.
To engineer the edits in gliadins, a 20-mer guideRNA (gRNA) plus the Cas9
endonuclease gene is introduced in immature wheat embryos. The gRNA directs the
Cas9 nuclease protein to the target site within the gene of interest, where the nuclease
generates a double-strand DNA break, three bases upstream of a specific protospacer
adjacent motif (PAM). In plants, repair by non-homologous end joining (NHEJ) can
result in insertion/deletions (indels) and other mutations creating potentially beneficial
phenotypes [15,16], and plants with beneficial mutations are selected from the
regenerated plants. Introducing a construct in wheat through genetic transformation
was not easy, but Ishida et al. [17] achieved a breakthrough using the US soft white
wheat variety, Fielder, employing a supervirulent Agrobacterium tumefaciens strain.
The introduced construct can be removed in crosses afterwards, simply be selecting
construct-free offspring.
An alternative approach for stable transformation is to use transient expression of the
CRISPR/Cas9 DNA and the guide RNA. This has been achieved in wheat [18],
although it remains to be seen if the efficiency is high enough to edit the multiple
gluten targets in one cell; this strategy requires very active promotors.
ddPCR screening
Random mutagenesis, as well as targeted gene editing with CRISPR/Cas9 of gluten
gene families will produce offspring with a mosaic of deleted, modified, and
unchanged genes (Jouanin et al., submitted). Hence, methods to screen the offspring,
as well as the lines from subsequent crosses, are essential. Strategies that aim at
producing safe gluten genes without epitopes would require more sophisticated
technologies than strategies aiming at wheat with all gluten genes deleted. Exome
capture methods have been developed, when combined with next generation (Illumina)
sequencing, to provide sensitivity at reasonable costs when hundreds of lines need to
be analysed (see below). However, there is still ample need for new and faster methods
for the pre-screening phase, when thousands of seeds may need to be screened.
Droplet digital PCR (ddPCR) could be one of the techniques used for this. ddPCR
enables high-throughput screening of copy number variation (CNV), and Jouanin et al.
(in preparation) have applied it successfully for detecting the number of α-gliadins.
SNP markers
Further, marker technology is a helpful tool in identifying and following haplotypes
through crosses and might be useful to follow variant alleles. Much progress has been
made in this field, thanks to the combination of next generation sequencing technology
for cheap (re)sequencing with the technology for high-throughput genotyping using
SNP markers. For example, Rimbert et al. [19] performed whole-genome resequencing
of eight bread wheat lines. They identified 3.3 million SNPs, 49% on the B-genome,
41% on the A-genome and 10% on the D-genome. The TaBW280K high-throughput
genotyping array contains 280,226 of these SNPs, of which 69% can be efficiently
scored, half of them showing a diploid-like clustering. This was, for example, used to
produce a dense genetic map of Chinese Spring x Renan, comprising 83,721 markers.
Capture sequencing
Although these dense SNP arrays will allow to uniquely tag haplotypes in the wheat
germplasm, it will not be possible to differentiate newly made CRISPR/Cas9-edited
and coeliac-safe gliadin genes from immunogenic gliadin genes. Resequencing the
whole genome of a wheat variety is still a very laborious task. Therefore, we recently
developed a DNA capture system, GlutEnSeq, that enables specifically capturing the
complete set of gluten genes (including all polymorphisms and modifications) in a
variety, an accession, or a progeny plant, after which this set can be sequenced and
analysed in detail (Jouanin et al. submitted). The GlutEnSeq system was based on the
REnSeq system (for Resistance genes Enrichment and Sequencing [20]) for capturing
and sequencing disease resistance genes and alleles in plants.
EU regulation of gene editing
Recently, the European Court of Justice ruled that, according to the text of Directive
2001/18/EC, all mutations artificially induced by gene editing (e.g. by CRISPR/Cas
technology) are GM, also if they are identical to mutations that could be induced by
random mutagenesis, or already exist elsewhere in nature. This implies that it may
require 10-100 M€ to perform all required safety tests for hypo-immunogenic wheat in
which gluten genes have been deleted and/or epitopes have been modified. Thus, it is
expected that safe or hypoimmunogenic wheat will not be brought on the EU market,
in contrast to the situation in the US and in other countries where the safety of the end
product of a technology, and not the technology itself, is considered [21].
Mutation breeding
Mutation breeding using gamma-irradiation or EMS, which causes random mutations,
is not regulated in the EU. EMS has been used for mutagenesis for many decades, also
in wheat. Krasileva et al. [22] analysed a collection of 2735 EMS-mutated bread wheat
and durum wheat lines using an 84-Mb exome capture assay followed by sequencing. They identified more than 6 million mutations in the hexaploid Cadenza background,
and more than 4 million in the tetraploid Kronos-derived lines. The lines had on
average 2705 (in Kronos) and 5351 (in Cadenza) mutations, equalling 35-40 mutations
per kb. Even at this high number of mutations, it would be very difficult to identify
lines in which many gluten genes would be mutated simultaneously, in the desired way
towards coeliac-safety. Combining the mutations from various lines by crossing may
be an option, followed by backcrossing to Cadenza or Kronos to remove most of the
other mutations, but identification of the mutants at the DNA sequence level is still
required.
The statistics become more attractive when one does not need to mutate all gluten
genes separately, but when a gene that influences their expression could be targeted.
Moehs et al. [23] developed a decreased-gluten wheat using EMS-mutated alleles of
the wheat homolog of lys3a in barley. For this they screened >10,000 M2 plants and
found as many as 350 mutations in the three wheat homoeologs of lys3a, a prolaminbox
binding factor that that leads to a higher content of free lysine in barley as a
consequence of lower prolamin synthesis. By combining three mutations they already
achieved 50-60% reduction in gliadin and LMW-glutenin accumulation. The authors
work for Arcadia Biosciences, California, USA, who develop and market varieties
with health benefits (lower gluten or slow-degrading starch). The approach is based
upon random mutagenesis methods in combination with high-throughput screening,
and then combining the mutations by crossing.
Wheat with coeliac-safe gluten or without gluten
Gliadin and glutenin genes without immunogenic epitopes do exist, but are rare and
always accompanied by many gluten genes with one or more epitopes. Wheat varieties
that are safe for coeliacs do not exist naturally and cannot be produced by conventional
breeding alone [6], because there are too many genes involved (too few of which have
coeliac-safe alleles) which are clustered on different chromosomes in each of the three
genomes in such a way that recombination is not effective to combine only the safe
alleles. Deleting blocks of genes with traditional mutagenesis leads to the loss of more
and different genes, which affects the fitness of the resulting plants. Deletion lines, e.g.
in Chinese Spring, in which parts of chromosome arms with gluten loci have been
eliminated by mutagenesis, are useful research tools, but usually not considered
suitable material for a breeding program.
An exception to this rule is the Ultra Low Gluten variety produced in diploid barley.
For this, gamma-irradiated accession Risø 56 (lacking B-hordeins), Risø 1508 (lacking
C-hordeins) and R118 (an Ethiopian derived line which lacks D-hordeins) have been
intercrossed to produce ULG 2.0, which has a hordein (gluten) content below 5 ppm,
thus allowing it to be classified as ‘gluten-free’ [24]. Subsequent crossing and
selection with commercial varieties resulted in a variety with good malting and
brewing characteristics, marketed as ‘Kebari’ and currently used for the production of
gluten-free beer in Germany. Interestingly, in several countries the products made with this variety cannot legally be marketed as gluten-free, solely because of the fact that it
contains barley.
Conclusion
Comparing the different strategies for mutagenesis, the targeted approach using the
CRISPR/Cas technology is promising and involves a relatively simple and directed
detection protocol for adequate analysis of the modified wheat lines: ddPCR followed
by capture sequencing. Further, towards the end of the development of gene-edited
coeliac-safe wheat lines, proteomics methods are needed to precisely characterise the
protein composition of the grains to fully confirm the desired gene editing.
Acknowledgements
The research mentioned was partially funded by FP7-PEOPLE-2013-ITN, EID Maximising
the potential of CROP researchers (Max-CROP), grant no. 607178. It does not
necessarily reflect the views of the European Commission and in no way anticipates
the Commission’s future policy in this area.
References
1. Van Herpen TWJM, Goryunova SV, Van der Schoot J, et al. Alpha-gliadin genes
from the A, B and D genomes of wheat contain different sets of celiac disease
epitopes. BMC Genomics 2006; 7: 1.
2. Goryunova SV, Salentijn EMJ, Chikida NN, et al. Expansion of the gamma-gliadin
gene family in Aegilops and Triticum. BMC Evol Biol 2012; 12: 215.
3. International Wheat Genome Sequencing Consortium (IWGSC). Shifting the limits
in wheat research and breeding using a fully annotated reference genome. Science
2018; 361: eaar7191.
4. Sollid LM, Qiao SW, Anderson RP, et al. Nomenclature and listing of celiac
disease relevant gluten T-cell epitopes restricted by HLA-DQ molecules.
Immunogenetics 2012; 64: 455-460.
5. Shewry PR, Tatham AS. Improving wheat to remove coeliac epitopes but retain
functionality. J Cereal Sci 2016; 67: 12-21.
6. Jouanin A, Gilissen LJWJ, Boyd LA, et al. Food processing and breeding strategies
for coeliac-safe and healthy wheat products. Food Res Int 2018; 110: 11-21.
7. Jia J, Zhao S, Kong X, et al. Aegilops tauschii draft genome sequence reveals a
gene repertoire for wheat adaptation. Nature 2013; 496: 91-95.
8. Luo MC, Gu YQ, You FM, et al. A 4-gigabase physical map unlocks the structure
and evolution of the complex genome of Aegilops tauschii, the wheat D-genome
progenitor. Proc Natl Acad Sci USA 2013, 110: 7940-7945.
9. Kumar A, Seetan R, Mergoum M. et al. Radiation hybrid maps of the D-genome of
Aegilops tauschii and their application in sequence assembly of large and complex
plant genomes. BMC Genomics 2015, 16: 800.
10. Dong L, Huo N, Wang Y, et al. Rapid evolutionary dynamics in a 2.8-Mb
chromosomal region containing multiple prolamin and resistance gene families in
Aegilops tauschii. Plant J 2016; 87: 495-506.
11. Huo N, Zhang S, Zhu T, et al. Gene duplication and evolution dynamics in the
homeologous regions harboring multiple prolamin and resistance gene families in
hexaploid wheat. Front Plant Sci. 2018; 9: 673.
12. Huo N, Zhu T, Altenbach S, et al. Dynamic evolution of α-gliadin prolamin gene
family in homeologous genomes of hexaploid wheat. Sci Rep 2018; 8: 5181.
13. Clavijo BJ, Venturini L, Schudoma C, et al. An improved assembly and annotation
of the allohexaploid wheat genome identifies complete families of agronomic
genes and provides genomic evidence for chromosomal translocations. Genome
Res 2017; 27: 885-896.
14. Sánchez‐León S, Gil‐Humanes J, Ozuna CV, et al. Low‐gluten, non‐transgenic
wheat engineered with CRISPR/Cas9. Plant Biotechnol J 2018; 16: 902-910.
15. Belhaj K, Chaparro-Garcia A, Kamoun S, et al. Plant genome editing made easy:
targeted mutagenesis in model and crop plants using the CRISPR/Cas system.
Plant Methods 2013; 9: 39.
16. Van de Wiel CCM, Schaart JG, Lotz LAP, et al. New traits in crops produced by
genome editing techniques based on deletions. Plant Biotechnol Rep 2017; 11: 1-8.
17. Ishida Y, Tsunashima M, Hiei Y, Komari T. Wheat (Triticum aestivum L.)
transformation using immature embryos. In: Wang K. (ed) Agrobacterium
Protocols. Methods in Molecular Biology, 3rd Edn., Springer, New York, NY,
2015; pp. 189-198.
18. Zhang Y, Liang Z, Zong Y, et al. Efficient and transgene-free genome editing in
wheat through transient expression of CRISPR/Cas9 DNA or RNA. Nat Commun
2016; 7:12617.
19. Rimbert H, Darrier B, Navarro J, et al. High throughput SNP discovery and
genotyping in hexaploid wheat. PLoS One 2018; 13:e0186329.
20. Jupe F, Witek K, Verweij W, et al. Resistance gene enrichment sequencing
(RenSeq) enables reannotation of the NB�LRR gene family from sequenced plant
genomes and rapid mapping of resistance loci in segregating populations. Plant J
2013; 76: 530-544.
21. Jouanin A, Boyd LA, Visser RGF, et al. Development of wheat with
hypoimmunogenic gluten obstructed by the gene editing policy in Europe. Front
Plant Sci 2018; 9: 1523.
22. Krasileva KV, Vasquez-Gross HA, Howell T, et al. Uncovering hidden variation in
polyploid wheat. Proc Natl Acad Sci USA 2017; 114: E913-E921.
23. Moehs CP, Austill WJ, Holm AP, et al. Development of decreased-gluten wheat
enabled by determination of the genetic basis of lys3a barley. Plant Physiol 2019;
https://doi.org/10.1104/pp.18.00771.
24. Tanner, G.J., Blundell, M.J., Colgrave, M.L., Howitt, C.A. Creation of the first
ultra-low gluten barley (Hordeum vulgare L.) for coeliac and gluten-intolerant
populations. Plant Biotechnol J 2016; 14: 1139-1150.
Statements on current developments
concerning gluten analysis, clinical and legal
aspects
Gluten-free diet & irritable bowel syndrome
Ombretta Polenghi, Virna Cerne
Dr. Schär R&D Centre, Trieste, Italy
Irritable bowel syndrome
Irritable bowel syndrome (IBS) is a common gastrointestinal disorder characterised by
abdominal pain, discomfort and alteration in bowel habits. IBS is also responsible for
psychological disorders like anxiety, depression and a general diminishing in quality
of life [1]. This disorder affects approximately 4-20% of western population.
IBS is connected to coeliac disease (CD) by presence of similar symptoms (diarrhoea,
nausea, abdominal pain), which causes, in some instances, CD to be misdiagnosed for
IBS. Non-celiac gluten sensitivity (NCGS) is characterised by intestinal and extraintestinal
symptoms related to the ingestion of gluten/wheat - containing food, in
subjects that are not affected by either CD or wheat allergy [2].
NCGS was described recently, after some cases of patients with symptoms very
similar to CD and IBS, but without abnormalities at intestinal cells, and whose
symptoms improved after an adherence to a gluten-free diet (GDF).
Fig. 1 shows the overlap between IBS, NCGS and CD symptoms. Symptoms are
shared between CD, IBS and NCGS, and all three conditions seem to have a benefit
from a gluten-free diet.
Causal agents for IBS are still under study and there are many hypothesis. Even if
sugar components are described as the most possible cause, Aziz et al. ^$ et al [1]
suggested also other components in wheat as responsible for IBS. Beside Fermentable
Oligosaccharides, Disaccharides, Monosaccharides And Polyols (FODMAPs), which
are short-chain carbohydrates causing gas production and osmotic diarrhoea, there are:
i) wheat germ agglutinin, which are known to stimulate the pro-inflammatory
cytokines; ii) amylase trypsin inhibitors (ATIs), which can stimulate the innate
immune response, activate the TLR4, and increase T-cell adaptive response and iii)
gluten that stimulates the innate immune response, inducing the proliferation of
monocytes, the expression of TLRs and INF-γ and altering intestinal permeability [3].
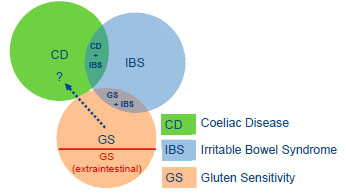
Figure 1. Overlapping between IBS, NCGS and CD [4].
FODMAPs and IBS
Therapy for IBS has not been discovered, while it is known that some kinds of foods
trigger IBS symptoms, researchers are still looking for an effective way to treat IBS.
Diet, in some cases, can be helpful; in particular, a low FODMAPs diet can reduce
symptoms. Already in 2006 research examined the role of FODMAPs in IBS. One
study showed a symptomatic improvement in 74% on a low-fructose/fructan diet [5].
Since then, several randomised control trials have demonstrated the efficacy of the low
FODMAPs diet and probable mechanisms [6].
Halmos et al. [7] have studied the effect of a FODMAPs-free diet in people with IBS,
in comparison with a high FODMAPs diet. Low FODMAPs diet was consumed for 21
days by 30 patients with IBS and 8 healthy individuals, followed by consumption of a
typical Australian diet, with a daily content of 4,4g oligosaccharides and 2,6g polyols,
for other 21 days. Depending on the type of diet, differences in symptoms were
reported. In IBS patients treated with low FODMAPs diet the results (Fig. 2) indicated
a clear decrease in pain and disturbs linked to the IBS, including low bloating (A) and
abdominal pain (B), a reduced faecal frequency in case of IBS-D (with diarrhoea) in D
graph, less dissatisfaction of stool consistency in IBS-C (with constipation) in E.
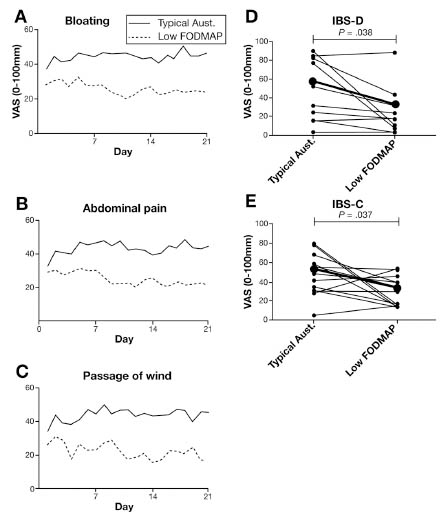
Figure 2. Mean of symptoms of IBS during a typical Australian diet and low
FODMAPs diet according to Halmos et al. (2014) [7].
Relation between Gluten and IBS
Research recently has focused also on the role of a gluten-free diet in IBS. The benefit
of a gluten-free diet in patients especially with IBS- D (and also IBS –M) has been
shown in different studies.
A clinical study demonstrated that GFD can improve symptoms in IBS patients [8]. A
GFD was followed by a group of IBS patients with different type of symptoms
(diarrhoea-dominant IBS-D and mixed type IBS-M). Among these, 34% were
responder to GFD, feeling a symptoms relief (Fig. 3).

Figure 3. Clinical study on 35 IBS patients supplied with GFD.
In a prospective study of 41 patients with IBS-D all participants were placed on a six
week gluten-free diet following advice by a dietitian, at six-week follow up. A GFD
had significantly reduced “IBS Symptom Severity Scores” in 71% of the cohort and
the mean total IBS symptom Severity Scores decreased from 286 to 131 points [9].
Even if the trigger of IBS is still not clear, in many studies it is reported that the GFD
brings to symptoms relief. This is probably due to low concentration of FODMAPs,
agglutinins, ATIs and gluten in the gluten-free foods.
Conclusions
IBS is a disorder connected with GS and CD. Its pathophysiology mechanism is not
clear and causal agents are still not identified. So far, GFD is a possible therapy, not
only for CD, but also for some IBS and NCGS patients. Further studies about IBS,
diet, and role of wheat in IBS are necessary for a better understanding of this
syndrome, as well as for the identification of all the critical elements that can trigger
symptoms and for the design of dedicated gluten-free food solutions.
References
1. Aziz I, Trott N, Briggs R, et al. Efficacy of a gluten-free diet in subjects with
irritable bowel syndrome-diarrhea unaware of their HLA-DQ2/8 genotype. Clin
Gastroenterol Hepatol 2016; 14: 696-703.e1.
2. Catassi C, Elli L, Bonaz B, et al. Diagnosis of non-celiac gluten sensitivity
(NCGS): The salerno experts’criteria. Nutrients 2015; 7:4966-4977.
3. Aziz I, Hadjivassiliou M, Sanders DS. The spectrum of noncoeliac gluten
sensitivity. Nat Rev Gastroenterol Hepatol 2015; 12(9): 516-626.
4. Verdu EF, Armstrong D, Murray JA. Between celiac disease and irritable bowel
syndrome: The “no man’s land” of gluten sensitivity. Am J Gastroenterol 2009;
104(6): 1587-1594.
5. Shepherd SJ, Gibson PR. Fructose malabsorption and symptome of irritable bowel
syndrome: Guidelines for effective dietary management. J Am Diet Assoc 2006;
106(10):1631-1639.
6. Trott N, Aziz I, Sanders D. Dietary Treatment in IBS. Progress, possibilities and
patient preference. Gastroenterol Hepatol 2017; 8(8): 552-554.
7. Halmos EP, Power VA, Shepherd SJ, et al. A diet low in FODMAPs reduces
symptoms of irritable bowel syndrome. Gastroenterology 2014; 146(1): 67-75.
8. El-Salhy M, Hatlebakk JG, Gilja OH, et al. The relation between celiac disease,
nonceliac gluten sensitivity and irritable bowel syndrome. Nutrition Journal 2015;
14: 92.
9. Barmeyer C, Schumann M, Meyer T, et al. Long-term response to gluten-free diet
as evidence for non-celiac wheat sensitivity in one third of patients with diarrheadominant
and mixed-type irritable bowel syndrome. Int J Colorectal Dis 2017; 32:
29-39.
On deamidation and hydrolysis of wheat gluten
Johan De Meester
Cargill R&D Centre Europe, Vilvoorde, Belgium
Introduction
Non enzymatic deamidation is a post-modification event that has been observed and
characterised in a wide variety of proteins playing a role in several diseases, and is
believed to relate to aging effects acting as a molecular clock [1,2]. It has been shown
to regulate some time dependent biological processes for the determination of the age
of e.g. bones [3]. There are many reports on deamidation under physiological
conditions in proteins of biological significance. Deamidation occurs in vitro and in
vivo, and the rates of deamidation depend on multiple factors, including the primary
sequences and three-dimensional structure, pH, temperature, and buffer salts in the
solutions [4].
Under mildly alkaline conditions, asparagine deamidation starts with the nucleophilic
attack of the peptide nitrogen on the side chain carbonyl of the following peptide
group leading to the formation of an asymmetric five-membered succinimide ring
intermediate that is quickly hydrolysed into D,L-aspartate and D,L-isoaspartate in a
ratio of approximately 1:3 [5-8]. The latter results in a -shift in the primary sequence.
Deamidation of glutamine is as much as ten times slower because it is thermodynamically
less favourable to form a six-membered glutarimide ring [9-11]. As a free
amino acid, or as the N-terminal residue of a peptide or protein, glutamine deamidates
readily to form pyroglutamic acid (5-oxoproline). The reaction proceeds via
nucleophilic attack of the α-amino group on the side-chain amide to form a γ-lactam
with the elimination of ammonia from the side-chain. At low pH (< 2), direct
hydrolysis of the side chain amide generates aspartate as the sole product without the
formation of a cyclic imide intermediate. Deamidation causes an increase in the mass
of 0.984 Da due to replacement of an amide into a carboxylic group.
The purpose of this contribution is to highlight protein chemical insights on
deamidation and hydrolysis of wheat gluten and oilseeds proteins showing that
chemical deamidation is not at random process but determined by the amino acid
composition and sequence of the protein source. Deamidation is one of the major
degradation process of proteins and peptides. Caution should be taken when storing
and preparing samples for analysis.
Deamidation in food proteins
Major cereal and oilseed proteins contain non-essential amino acids between 55 to 69
% with cereal flours covering the higher end (57-69 %) [12]. Oilseed proteins are containing higher levels of arginine and lysine. Glutamine and asparagine are making
up 23-43 % of the total amount of amino acids. Table 1 is summarizing some relevant
data on glutamine and asparagine levels of different protein sources. The ratio between
glutamine to asparagine in cereals is quite high (for wheat derived products 7 or higher
even higher than 10 for wheat gluten) compared to the ratio found in other protein
sources (below 3). Another feature is the level of amidation : in cereal products levels
between 80 to 96 % are found, while in oilseed proteins the level of amidation is about
50 to 60 %.
Table 1. Glutamine and aspartic acid parameters from major cereals and oilseeds
crops
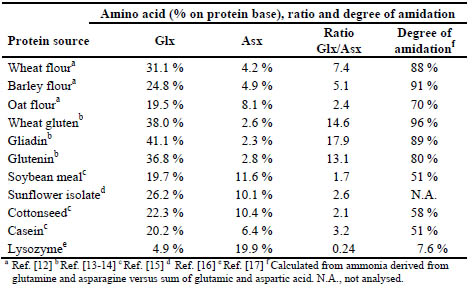
The amino acid composition of wheat gluten is characterised by a high glutamic acid
content, a relatively high proline content, a low amount of basic amino acids (lysine,
arginine, histidine), a high degree of amidation nearly equivalent to the aspartic and
glutamic acid content and a remarkable cysteine content. Aspartic acid content in
wheat gluten is lower compared to the level in wheat flour. The mean percentage of
(glutamic + aspartic) acids in the amide form is 82 for glutenin and 92 for gliadin
[18,19]. More recent determinations on amino acid composition on gluten, gliadin and
glutenin [20] revealed that the ratio of glutamine to asparagine was between 10 to 11.
A high degree of amidation increase the hydrogen bonds interactions and decrease
solubility. Lásztity [21] showed that the relation between the degree of amidation and
the relaxation time of gluten showed an optimum at 88 % of amidation. Deamidation
introduces a negative charge at the reaction site and can also lead to structural
isomerisation especially around aspartyl residues containing amino acids with low
steric hindrance. The extent and rate of deamidation is depending on the ratio of glutamine and asparagine and the degree of amidation in the native protein. pH is also
very important, not only to the extent of deamidation but also to the mechanism of the
reaction. Zhang et al. [22] compared the kinetics of deamidation of soy protein isolate
with that of lysozyme. The deamidation rate of lysozyme was higher compared to that
of soy protein isolate which was explained by a much higher amount of asparagine
compared to glutamine in lysozyme. On either side of aspartic acid peptide bonds may
be cleaved at a rate at least 100 times greater than other bonds under carefully
controlled conditions in dilute acid. Free aspartic acid was observed 30 minutes after
the start of acidic deamidation of soy protein isolate which increased over time [23].
Deamidation with food proteins at large scale is mainly conducted by using acids [24].
Deamidation and hydrolysis of wheat gluten
A number of authors [25-28] have established that deamidation and hydrolysis of
wheat gluten go hand in hand. Finley [27] prepared suspensions of 5-10 % gluten with
hydrochloric acid of different strengths between 0.025 to 1 N HCl and reacted them
during 30 minutes at 50, 75 and 95 °C. Degree of hydrolysis and deamidation were
monitored. Peptide hydrolysis requiring much harsher reaction conditions (6 N HCl at
110 °C during 24 hours) as for amino acid analysis and can be viewed as hydrolysis of
a secondary amide, while complete deamidation only requiring treatment with 2 N
HCl at 110 °C during two hours. The sum of deamidation (= hydrolysis of primary
amide) and hydrolysis are expressed as hydrolysis of amide groups and is represented
in Fig. 1.
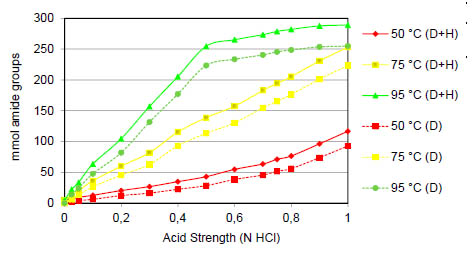
Figure 1. The effect of acid strength on the sum of deamidation (D) and hydrolysis (H)
of gluten suspensions at 50, 75 and 95 °C (data replotted from Finley, 1975 [27]).
As with most reactions, higher temperatures will increase the rate of the deamidation
reaction. Deamidation is for each treatment the main contributor in amide hydrolysis
compared to peptide hydrolysis although at 95 °C the contribution is remarkably
lower.
The contribution of hydrolysis is increasing only slightly as deamidation is progressing
to a much larger extent. In Fig 2 the Finley data has been replotted for each data point
and revealing that at each temperature the increase of deamidation and hydrolysis
follows the same pattern. Up to 25 % of deamidation the % DH (degree of hydrolysis)
is limited to 2.5-3 % while by levels of deamidation above 25 %, the hydrolysis goes
to a pace with is only one fourth of the initial level. This is explained by changes in the
higher-order structure in the protein [29]. A similar behaviour is observed by
deamidation of soy protein isolate although at 36 % deamidation a 4.5 % DH is
already obtained. This might be explained by the presence of more aspartic acid
residues in the soy protein at start which are most likely less amidated compared to
wheat gluten.
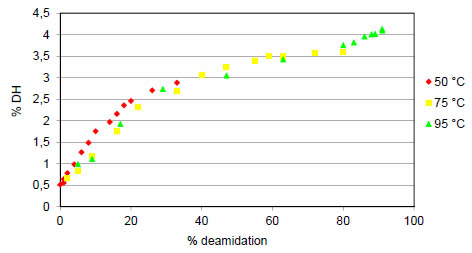
Figure 2. Recalculation and representation of the data from Fig. 1 and [27]. For each
data point the degree of hydrolysis of the peptide bond is expressed as a function of the
degree of deamidation.
Effect of deamidation on protein properties
Deamidation also was reported to increase exposed hydrophobicity of gluten induced
by a conformational change and, subsequently, to increase surface activity [29]. The
functional properties as emulsifying capacity and stability were increased greatly. A
number of studies investigated the molecular mechanism for protein structural
destabilisation upon deamidation. For example, the deamidation treatment has direct implications for charge density and, in turn, affects electrostatic interactions the
protein may undergo by interacting with water or upon self-assembly [9,27,30]. This
role of electrostatics to deamidation induced disruption of protein structure was
supported by further observations on wheat gluten: both acetic acid and HCl induced
deamidation had substantial consequences for the secondary structure of wheat gluten.
It was thus postulated that strong deamidation induced protein unfolding as a result of
electrostatic repulsion [31]. Acetic acid induced deamidation of wheat gluten was
further found to inhibit SDS-stable aggregate formation whilst largely retaining its
ability to form disulphide bonds [31]. The rate of the deamidation reaction has been
found to depend on primary sequence and pH under which the reaction takes place, but
was independent of ionic strength for model peptides [4,7,8,32], soy protein and egg
white lysozyme [22].
As glutamic and aspartic acid residues are the main constituents of the β-turn form, the
increase of β-turn might be due to the fact that more glutamic and aspartic acid
residues were exposed as the degree of deamidation increased. As a result of increased β-turn in modified samples, the charge density and electrostatic repulsion of protein
consequently became stronger after deamidation with HCl, facilitating unfolding of the
protein conformation and protein−water interactions and improving the susceptibility
of wheat gluten to enzymatic hydrolysis [33,34]. The increase of charge and decrease
of β-sheet make the proteins more flexible after deamidation, especially N-terminal
domain which was rich in glutamine residues. In comparison, the slight increase of helix which appeared mostly in C-terminal domain [35] means that C-terminal
domain is more resistant to changes induced by deamidation. Qiu et al. [34] reported
that wheat gliadin deamidated by HCl exhibited an increase of -sheet while a
decrease of the -helix was observed.
Deamidation using four different organic acids providing a similar degree of
deamidation with concomitant hydrolysis at the same levels as previously published.
Wang et al. [36] showed that the solubility and emulsifying property of deamidated
wheat glutens were significantly improved, and the essential amino acid contents were
enhanced. Circular dichroism and Fourier transform infrared spectroscopy showed
there was a decrease in -sheet and a slight increase in random coil of deamidated
wheat glutens. Deamidation of wheat gluten by succinic acid [33] was found to be
more efficient than that with citric acid, although wheat gluten treated with succinic
acid exhibited less improvement in the foaming capacity and stability and experienced
inhibition in the emulsification activity compared with the gluten treated with citric
acid. Wheat gluten deamidated with citric acid exhibited more flexible protein
molecules, greater changes in the tertiary and secondary structures. Moreover, succinic
acid deamidation-induced modification resulted in little change in molecular weight
and secondary structure of the protein. Thus, succinic acid could facilitate unfolding
protein conformation. Moisture-heating (121 °C during 10 min) in the deamidation
pre-treatment decreased the susceptibility of wheat gluten to enzymatic hydrolysis and
the peptide factions of ≤ 3000 Da in the hydrolysates due to the formation of larger
molecule weight aggregates.
Conclusion
Industrially deamidated wheat glutens are manufactured using acids providing
products with a degree of deamidation between 15 to 60 % [24]. However, although
the same degree of deamidation (60 %) and hydrolysis was obtained using different
organic acids, differences in the release of free amino acids was evidenced [37]
following a complementary enzymatic hydrolysis during 48 hours using pancreatin.
Most likely due to a different secondary and tertiary structure in deamidated wheat
gluten brought by deploying different organic acids leading to the formation of
carboxylic groups and reduction of hydrogen bonds in a different high order structure.
Based on the faster deamidation at neutral and alkaline deamidation it is important to
take into account non enzymatic deamidation that easily occurs during sample
preparation using pancreatin at pH of 9. The impact on proteomic sample preparation
has not been systematically investigated. Mild acidic conditions eliminating
deamidation during proteolysis could improve conclusions on deamidated wheat
gluten when peptides are investigated. Recently, similar recommendations were
published for investigating the complexity of allergic diseases that approaches
combining accurate product characterisation of deamidated wheat gluten, in vitro and
animal models studies need to be considered [24].
References
1. Robinson NE, Robinson AB. Mechanisms of reactions involving Asn and Gln. In:
Robinson NE, Robinson AB, (eds.): Molecular Clocks, Cave Junction, Althouse,
2004. http://www.deamidation.org/MolecularClocks.pdf.
2. Robinson NE, Robinson AB. Amide molecular clocks in Drosophila proteins:
Potential regulators of aging and other processes. Mechanisms of Ageing and
Development 2004; 125: 259-267.
3. Simpson J. The effects of demineralisation and sampling point variability on the
measurement of glutamine deamidation in type I collagen extracted from bone. J
Archaeol Sci 2016; 69: 29-38.
4. Tyler-Cross R, Schirch V. Effects of amino acid sequence, buffers, and ionic
strength on the rate and mechanism of deamidation of asparagine residues in small
peptides. J Biol Chem 1991; 266: 22549-22556.
5. Geiger T, Clarke S. Deamidation, isomerization, and racemization at asparaginyl
and aspartyl residues in peptides. Succinimide-linked reactions that contribute to
protein degradation. J Biol Chem 1987; 262: 785-794.
6. Stephenson RC, Clarke S. Succinimide formation from aspartyl and asparaginyl
peptides as a model for the spontaneous degradation of proteins. J Biol Chem 1989;
264: 6164-6170.
7. Patel, K.; Borchardt, R. T. Chemical pathways of peptide degradation. II. Kinetics
of deamidation of an asparaginyl residue in a model hexapeptide. Pharm Res 1990;
7: 703-711.
8. Patel, K.; Borchardt, R. T. Chemical pathways of peptide degradation. III. Effect of
primary sequence on the pathways of deamidation of asparaginyl residues in
hexapeptides. Pharm. Res. 1990; 7: 787-793.
9. Wright HT. Nonenzymatic deamidation of asparaginyl and glutaminyl residues in
proteins. Crit Rev Biochem Mol Biol 1991; 26: 1-52.
10. Robinson NE, Robinson ZW, Robinson BR, et al. Structure-dependent nonenzymatic
deamidation of glutaminyl and asparaginyl pentapeptides. J Peptide Res
2004; 63: 426-436.
11. Robinson NE, Robinson AB. Prediction of primary structure deamidation rates of
asparaginyl and glutaminyl peptides through steric and catalytic effects. J Peptide
Res 2004; 63: 437-448.
12. Koehler P, Wieser H. Chemistry of Cereal Grains. In: Gobbetti M, Gänzle M
(eds.), Handbook on Sourdough Biotechnology, Springer Science&Business Media
New York 2013. Chapter 2: pp. 11-44.
13. Wu YV, Dimler RJ. Hydrogen ion equilibria of wheat glutenin and gliadin. Arch
Biochem Biophys 1963; 103: 310-318.
14. Wu YV, Dimler RJ. Hydrogen ion equilibria of wheat gluten. Arch Biochem
Biophys 1963; 102: 203-237.
15. Shih FF. Deamidation studies on selected food proteins. JAOCS 1990; 67: 675-677.
16. Ivanova P, Chalova V, Koleva L, et al. Amino acid composition and solubility of
proteins isolated from sunflower meal produced in Bulgaria. Int Food Res J 2013;
20: 2995-3000.
17. Canfield R. The amino acid sequence of egg white lysozyme. J Biol Chem 1963;
238: 2698-2707.
18. Ewart JAD. Amino acid analyses of cereal flour proteins. J Sci Food Agric 1967;
18: 548-552.
19. Ewart JAD, Amino acid analyses of glutenins and gliadins. J Sci Food Agric 1967;
18: 111-116.
20. Rombouts I, Lamberts L, Celus I, et al. Wheat gluten amino acid composition
analysis by high-performance anion-exchange chromatography with integrated
amperometric detection. J Chrom 2009; 1216: 5557-5562.
21. Lásztity R. Correlation between the chemical structure and rheological properties
of gluten. Period Polytech, Chem Eng 1982; 26: 3-25.
22. Zhang J, Lee TC, Ho, CT. Comparative study on kinetics of nonenzymatic
deamidation of soy protein and egg white lysozyme J Agric Food Chem 1993; 41:
2286-2290.
23. Matsudomi N, Kato A, Kobayashi K. Conformation and functional properties of
acid-modified soy protein. Agric Biol Chem 1985; 49: 1251-1256.
24. Tranquet O, Larré C, Denery-Papini S. Allergic reactions to hydrolysed wheat
proteins: clinical aspects and molecular structures of the allergens involved. Crit
Rev Food Sci Nutr. 2018; 21: 1-10.
25. Vickery HB. The rate of hydrolysis of wheat gliadin. J Biol Chem 1922; 53: 495-512.
26. Vickery HB. A product of mild hydrolysis of wheat gliadin. J Biol Chem 1923; 56:
415-428.
27. Finley JW. Deamidated wheat gluten: a potential fortifier for fruit juices. J Food
Sci 1974; 40: 1283-1285.
28. Wu CH, Nakai S, Powrie WD. Preparation and properties of acid-solubilized
gluten. J Agric Food Chem 1976 24: 504-510.
29. Matsudomi N, Kato A, Kobayashi K. Conformation and surface properties of
deamidated gluten. Agric Biol Chem 1982; 46: 1583-1586.
30. Riha WE, Izzo HV, Zhang J, et al. Nonenzymatic deamidation of food proteins.
Crit Rev Food Sci Nutr 1996; 6: 225-255.
31. Liao L, Qiu CY, Liu TX, et al. Susceptibility of wheat gluten to enzymatic
hydrolysis following deamidation with acetic acid and sensory characteristics of
the resultant hydrolysates. J Cereal Sci 2010; 52: 395-403.
32. Robinson AB, Rudd C. Deamidation of glutaminyl and asparaginyl residues in
peptides and proteins. Curr Top Cell Regul 1974; 8: 247-295.
33. Liao L, Zhao M , Zhao, H. et al. Effect of succinic acid deamidation-induced
modification on wheat gluten. Front Chem Eng China 2009; 3: 386-392.
34. Qiu C, Sun W, Su G, et al. Comparison of the conformational and nutritional
changes of deamidated wheat gliadin by citric acid and hydrochloric acid. J Cereal
Sci 2014; 60: 143-150.
35. Wong BT, Zhai J, Hoffmann SV, et al. Conformational changes to deamidated
wheat gliadins and -casein upon adsorption to oil-water emulsion interfaces. Food
Hydrocoll 2012; 27: 91-101.
36. Wang Y, Gan J, Zhou Y, et al. Improving solubility and emulsifying property of
wheat gluten by deamidation with four different acids: Effect of replacement of
folded conformation by extended structure. Food Hydrocoll 2017; 72: 105-114.
37. Liao L, Wang Q, Zhao M. Investigation of the susceptibility of acid-deamidated
wheat gluten to in vitro enzymatic hydrolysis using Raman spectra and free amino
acid analysis. J Sci Food Agric 2012; 92: 1865-1873.
Perspectives and action plan of the PWG
Peter Koehler
Biotask AG, Esslingen, Germany
The Prolamin Working Group executive meeting and joint discussion held on 28
September 2018, led to the decisions and statements outlined below.
Action plan
I. Analytical
Efforts to find an alternative distributor for PWG-gliadin have finally been
successful. From 1st January, 2019, the material is available from the
Arbeitsgemeinschaft Getreidedeforschung e.V. (Association of Cereal
Research), Detmold, Germany.
Alternative reference material: The MoniQA initiative has extensively
characterised wheat cultivars from around the globe. A flour of a blend of five
wheat cultivars will shortly be available as a reference material for purchase.
However, the PWG considers flour not as a suitable reference material and
supports a protein sample as reference material. Further action is planned in this
field.
II. Clinical
To improve the visibility of the group, Open Access publications with scientific
opinions of the PWG on current issues of cereals and gluten will be made.
The PWG considers being a working group under the umbrella of the
International Society For The Study Of Celiac Disease
III. Members, Policy
Carmen Gianfrani is a new member of the group.
Martin Stern, Thomas Mothes and Rudolf Valenta left the group.
New group members have to be identified because some members will retire in
the next years.
This printed, citable book (print run: 250 copies with ISBN number) was made
possible by funding of Dr. SCHÄR GmbH/Srl, (Burgstall, BZ, Italy). It will be
distributed among leaders of opinion in gluten analysis and clinical medicine.
An electronic version can be downloaded free of charge from the PWG website
(http://www.wgpat.com).
Next meeting: 2019
We are very pleased to announce the venue for our meeting in 2019:
Urbino, Italy
Host:
Prof. Dr. Carlo Catassi
Università Politecnica delle Marche
Ancona, Italy
E-mail: c.catassi@staff.univpm.it
Time: 10 - 12 October 2019
Focus of the meeting:
Research on amylase-trypsin inhibitors (ATI)
The meeting will be limited to 55 participants and attendance is by
invitation only. Invitations will be sent by April 2019. Registration
deadline will be June 15, 2019.
Very special thanks to the host of this kind invitation!
|
|
List of Participants
GROUP MEMBERS
Prof. Dr. Fernando G. Chirdo
Universidad Nacional de La Plata
Facultad de Ciencias Exactas
Instituto de Estudios Immunologicos y
Fisiopatologicos - IIFP
Calle 47 y 115
(1900) LA PLATA, ARGENTINA
Phone: +54 221 423 5 333 (Int 45)
Fax: +54 221 422 6947
E-mail: fchirdo@biol.unlp.edu.ar
Prof. Dr. Paul J. Ciclitira
GSTT NHS Trust
Curve Business Hub, Unit 1
30B Wilds Rents
SE1 4QG LONDON
UNITED KINGDOM
Phone: +44 203 751 1104
Fax: +44 207 4033437
E-mail: pjcgastro@gmail.com
Prof. Dr. Carlo Catassi
(not attending), substituted by
Dr. Anil K. Verma
Università Politecnica delle Marche
Celiac Disease Research Laboratory
Dept of Pediatrics
Via Cadorna 5
60123 ANCONA, ITALY
Phone: +39 33 4292 2991
E-mail: anilkrvermaa@gmail.com
Prof. Dr. Conleth Feighery, MD
University of Dublin, Department of
Immunology, St. James’s Hospital
James’s Street
DUBLIN 8, IRELAND
Phone: +353 879969041
E-mail: con.feighery@tcd.ie
Prof. Dr. Peter Koehler
Biotask AG
Schelztorstraße 54-56
73728 ESSLINGEN, GERMANY
Phone: +49 711 31059068
Fax: +49 711 31059070
E-mail: peter.koehler@biotask.de
Prof. Dr. Knut Lundin
University of Oslo
Institute of Clinical Medicine
Postboks 1171, Blindern
0881 OSLO, NORWAY
Phone: +47 90980325
Fax: +47 23072410
E-mail: knut.lundin@medisin.uio.no
Prof. Dr. Thomas Mothes
Institut für Labormedizin der Universität
Leipzig
Liebigstraße 27
04103 LEIPZIG, GERMANY
Phone: +49 1577 3327893
Fax: +49 341 97 22373
E-mail: mothes@medizin.uni-leipzig.de
Dr. Katharina Scherf
Leibniz-Institut für Lebensmittel-
Systembiologie an der Technischen
Universität München
Lise Meitner-Straße 34
85354 FREISING, GERMANY
Phone: +49 8161712927
Fax: +49 8161712970
E-mail: k.scherf.leibniz-lsb@tum.de
Prof. Dr. Dr. Detlef Schuppan
I. Medizinische Klinik und Poliklinik
Universitätsmedizin der Johannes
Gutenberg-Universität Mainz
Institut für Translationale Medizin
Langenbeckstraße 1
55131 MAINZ, GERMANY
Phone: +49 6131 177355/177356/177104
Fax: +49 6131 177357
E-mail: detlef.schuppan@unimedizinmainz.de
Dr. René Smulders
Wageningen University & Research,
Plant Research
Droevendaalsesteeg 1
6708 PB WAGENINGEN,
THE NETHETRLANDS
Phone: +31 620298266
E-mail: rene.smulders@wur.nl
Dr. Olivier Tranquet
INRA
Rue de la Géraudière BP 71627
44316 NANTES CEDEX 3, FRANCE
Phone: +33 2 40675027
Fax: +33 240675025
E-mail: olivier.tranquet@inra.fr
Prof. Dr. Riccardo Troncone
Department of Pediatrics and European
Laboratory for the Investigation of
Food-induced Diseases
Department of Padiatrics
Via Pansini 5
80131 NAPLES, ITALY
Phone: +39 3483132274
Fax: +39 0817463116
E-mail: troncone@unina.it
Prof. Dr. Frits Koning
(not attending)
Leiden University Medical Centre, E3-Q
Department of Immunohaematology
and Bloodbank
Albinusdreef 2
2333 ZA LEIDEN, THE NETHERLANDS
Phone: +31 715 266673
Fax: +31 715 265267
E-mail: fkoning@lumc.nl
HOSTS
Ms. Pauline Titchener
Neogen Europe Ltd.
The Dairy School, Auchincruive, Ayr
KA6 5HU AYR, SCOTLAND,
UNITED KINGDOM
Phone: +44 1292 525 600
Fax: +44 1292 525 601
E-mail: p.titchener@neogeneurope.com
Ms. Sharon Forsythe
Neogen Europe Ltd.
The Dairy School, Auchincruive, Ayr
KA6 5HU AYR, SCOTLAND,
UNITED KINGDOM
Phone: +44 1292 525 600
Fax: +44 1292 525 601
E-mail: s.forsythe@neogeneurope.com
INVITED SPEAKERS
Dr. Manuel Spannagl
Helmholtz Zentrum München
Deutsches Forschungszentrum für
Gesundheit und Umwelt (GmbH)
Ingolstädter Landstraße 1
85764 NEUHERBERG, GERMANY
Phone: +49 89 31873948
Fax: +49 89 31872627
E-Mail: manuel.spannagl@helmholtzmuenchen.
de
GUESTS
Mrs. Sofia Beisel
Deutsche Zöliakiegesellschaft e.V.
Kupferstraße 36
70565 STUTTGART, GERMANY
Phone: +49 711 45998115
Fax: 49 711 45998150
E-mail: sofia.beisel@dzg-online.de
Mrs. Zsofía Birinyi
MTA Centre of Agricultural Research
Department of Applied Genomics
Brunszvik u 2
2462 MARTONVÁSÁR, HUNGARY
Phone: +36 225 69521
Fax: +36 225 69514
E-mail: birinyi.zsofia@agrar.mta.hu
Mr. Gregory Bouchaud
INRA
Rue de la Géraudière BP 71627
44316 NANTES, FRANCE
Phone: +33 2406 75142
Fax: +33 2406 75025
E-mail: gregory.bouchaud@inra.fr
Dr. Angel Cebolla-Ramirez
Biomedal, SL
Av. Américo Vespucino n° 5
- Bloque 4 - 1a planta -
41092 SEVILLA, SPAIN
Phone: +34 954 081276
Fax: +34 954 081279
E-mail: acebolla@biomedal.com
Dr. Virna Cerne
Dr. Schär SPA
Winkelau 9
30914 POSTAL, ITALY
Phone: +39 040 3755380
E-mail: arianna.grabbio@drschaer.com
Dr. Johan De Meester
Cargill R&D Centre Europe
Havenstraat 84
B-1800 VILVOORDE, BELGIUM
Phone: +32 225 70733
E-mail: Johan_De_Meester@cargill.com
Mr. Richard Fielder
Bio-Check (UK)
Spectrum House, Llys Edmund Prys,
St. Asaph Business Park
LL17 0JA ST. ASAPH,
UNITED KINGDOM
Phone: +44 1745 335165
Fax: +44 1745 582867
E-mail: richard@biocheck.uk.com
Dr. Gyöngyvér Gell
MTA Centre of Agricultural Research
Department of Applied Genomics
Brunszvik u 2
2462 MARTONVÁSÁR, HUNGARY
Phone: +36 225 69521
Fax: +36 225 69514
E-mail: gell.gyongyver@agrar.mta.hu
Mrs. Anna Gibert Casamada
SMAP Celíacs Catalunya
Independencia, 257
08026 BARCELONA, SPAIN
Phone: +34 934 121789
E-mail: eroger@celiacscatalunya.org
Dr. Thomas Grace
Bia Diagnostics/Elution Technologies
480 Hercules Dr.
5446 COLCHESTER, VT, USA
Phone: +1 802 540 0148
Fax: +1 8025400147
E-mail: thomasgrace@biadiagnostics.com
Mrs. Robin Grace
Bia Diagnostics/Elution Technologies
480 Hercules Dr.
5446 COLCHESTER, VT, USA
Phone: +1 802 540 0148
E-mail: robin@biadiagnostics.com
Dr. Reka Haraszi
Campden BRI
Station Road
GL556LD CHIPPING CAMPDEN,
UNITED KINGDOM
Phone: +44 1386842240
E-mail: reka.haraszi@campdenbri.co.uk
Ms. Barbara Lexhaller
Leibniz-Institut für Lebensmittel-
Systembiologie an der Technischen
Universität München
Lise Meitner-Straße 34
85354 FREISING, GERMANY
Phone: +49 8161712925
Fax: +49 8161712970
E-mail: b.lexhaller.leibniz-lsb@tum.de
Ms. Chiara Palladino
Romer Labs Division Holding GmbH
Erber Campus 1
3131 GETZERSDORF, AUSTRIA
Phone: +43 664 8847 2683
E-mail: chiara.palladino@romerlabs.com
Ms. Catherine Pidsley
R-Biopharm Rhone Ltd.
Block 10 Todd Campus, West Scotland
Science Park, Acre Road
G20 0XA GLASGOW,
UNITED KINGDOM
Phone: +44 141945 2942
Fax: 141945 2925
E-Mail: Catherine@r-biopharmrhone.com
Mrs. Ombretta Polenghi
Dr. Schär R&D Centre
c/o AREA Science Park
Padriciano, 99
34149 TRIESTE, ITALY
Phone: +39 040 3755381
E-mail: ombretta.polenghi@drschaer.com
Dr. Elena Quesada Hernández
Biomedal, SL
Av. Américo Vespucino n° 5
- Bloque 4 - 1a planta -
41092 SEVILLA, SPAIN
Phone: +34 954 081276
Fax: +34 954 081279
E-mail: elena.quesada@biomedal.com
Dr. Adrian Rogers
Romer Labs UK Ltd.
The Health Business and
Technical Park
WA74QX RUNCORN, CHESHIRE,
UNITED KINGDOM
Phone: +44 845519 0510
E-mail: adrian.rogers@romerlabs.com
Ms. Cristina Romero
Ingenasa
C/Hermanos García Noblejas, 39
28037 MADRID, SPAIN
Phone: +34 913 680501
E-mail: cromero@ingenasa.com
Dr. Annette Sauer
R-Biopharm AG
An der neuen Bergstraße 17
64297 DARMSTADT, GERMANY
Phone: +49 6151 81027291
Fax: +49 6151 81028099
E-mail: A.Sauer@r-biopharm.de
Ms. Eszter Schall
Budapest University of Technology and
Economics
Műegyetem rkp. 3
111 Budapest, Hungary
Phone: +36 2077 38835
E-mail: s.eszter@mail.bme.hu
Mr. Stefan Schmidt
R-Biopharm AG
An der neuen Bergstraße 17
64297 DARMSTADT, GERMANY
Phone: +49 151 29808524
Fax: +49 6151 810240
E-mail: st.schmidt@r-biopharm.de
Ms. Karoline Schreiber
Böcker Sauerteig GmbH & Co. KG
Ringstraße 55-57
32427 MINDEN, GERMANY
Phone: +49 571 837990
E-mail: karoline.schreiber@sauerteig.de
Dr. Juan Ignacio Serrano-Vela
Asociación de Celíacos y Sensibles Al
Gluten, Comunidad de Madrid
Calle Lanuza 19-bajo
28028 MADRID, SPAIN
Phone: +34 917 130147
Fax: +34 917 258059
E-mail:
nachoserrano@celiacosmadrid.org
Prof. Dr. Edurne Simón
University of the Basque Country
UPV/EHU
Paseo de la Universidad, 7
1006 VITORIA-GASTEIZ, SPAIN
Phone: +34 945 013069
Fax: +34 945 013014
E-mail: edurne.simon@ehu.eus
Dr. Heidi Urwin
Coeliac UK
3rd Floor, Apollo Centre, Desborough
Road
HP11 2QW HIGH WYCOMBE,
BUCKS, UNITED KINGDOM
Phone: +44 1494 796138
E-mail: Heidi.Urwin@coeliac.org.uk
Dr. Ángel Venteo
Ingenasa
C/Hermanos García Noblejas, 39
28037 MADRID, SPAIN
Phone: +34 913 680501
E-mail: aventeo@ingenasa.com
Ms. Clélia Villemin
INRA
Rue de la Géraudière BP 71627
44316 NANTES, FRANCE
Phone: +33 2406 75027
Fax: +33 2406 75025
E-mail: clelia.villemin@inra.fr
Mr. Niklas Weber
R-Biopharm AG
An der neuen Bergstraße 17
64297 DARMSTADT, GERMANY
Phone: +49 6151 81027222
Fax: +40 6151 81028099
E-mail: n.weber@r-biopharm.de
Dr. Paul Wehling
General Mills, Inc.
9000 Plymouth Ave N
55427 GOLDEN VALLEY, USA
Phone: +1 763 7644360
E-mail: paul.wehling@genmills.com
Dr. Thomas Weiss
R-Biopharm AG
An der neuen Bergstraße 17
64297 DARMSTADT, GERMANY
Phone: +49 6151 8102186
E-mail: t.weiss@r-biopharm.de
Mrs. Maren Wiese
Hermann Kröner GmbH
Lengericher Straße 158
49479 IBBENBÜREN, GERMANY
Phone: +49 5451 944712
E-mail: wiese@kroener-staerke.de
Impressum
Proceedings of the 32th Meeting
WORKING GROUP
on PROLAMIN ANALYSIS and TOXICITY
27 - 29 September 2018
Ayr, Scotland, UK
This work including all parts is subject to copyright. All rights are reserved and any
utilisation is only permitted under the provisions of the German Copyright Law.
Permissions for use must always be obtained from the publisher. This is in particular
valid for reproduction, translation, conversion to microfilm and for storage or
processing in electronic systems.
Scientific Organisation
Prof. Dr. Peter Koehler
biotask AG
Schelztorstraße 54-56, 73728 ESSLINGEN, GERMANY
Phone: +49 711 31059068; Fax: +49 711 31059070
E-mail: peter.koehler@biotask.de
Host
Pauline Titchener
Product Manager - Allergens and Speciation
European Headquarters of Neogen Corporation
The Dairy School, Auchincruive, AYR, KA6 5HU, SCOTLAND, UK
Phone: +44 1292 525 600; Fax: +44 1292 525 601
E-mail: p.titchener@neogeneurope.co.uk
Cover picture* and picture of participants:
Thomas Mothes
© Peter Koehler 2019
ISBN: 978-3-00-062148-2
|

