Proceedings of the 30th Meeting
Working Group on Prolamin Analysis
and Toxicity (PWG)
Edited by Peter Koehler
German Research Centre for Food Chemistry, Freising
Verlag Deutsche Forschungsanstalt für Lebensmittelchemie - 2016
Preface
In October 2014, I asked Cristina Rosell if she would be willing to host the 30th
meeting of the Working Group on Prolamin Analysis and Toxicity (PWG) and she
accepted with pleasure. She learned about the PWG meeting as a guest at the 2015
meeting in Tulln, Austria and started planning “her own” meeting. Together with her
colleague Maria Saneustaquio she organised the PWG meeting 2016 at the Hotel
Sercotel Sorolla Palace, Valencia, Spain from 22 to 24 September 2016. Cristina and
Maria were present during the entire meeting. As the chairman of the PWG, I assume
that going to Valencia at this weekend was like returning into summer. The PWG was
hosted by the Institute of Agrochemistry and Food Technology (IATA-CSIC) and the “Asociacón de Celíacos de la Communidad Valenciana”. The PWG, the invited
speakers, the participants from industry (cereal starch producers, producers of glutenfree
food, producers of kits for gluten analysis) and research institutes as well as the
delegates from European coeliac societies came together and had very interesting oneand-
a-half days of presentations, discussions and networking.
Analytical and clinical work in the field of coeliac disease and gluten done in the labs
of the PWG members as well as results of guests and invited speakers were presented
in 22 talks and intensely discussed at the meeting. In addition, one presentation was
focussed on regulatory aspects of gluten analysis and labelling. This was the highest
number of presentations at the PWG meeting during the last decade. A symposium on “Enzymatic Gluten Degradation” with two presentations of internationally recognised
experts highlighted the latest advances in the field of gluten-specific peptidases.
I would like to express my thanks to all participants of the meeting for their active
contributions and the discussions that resulted thereof. I am in particular grateful to
Maria Saneustaquio and Cristina Rosell from IATA-CSIC for their enthusiasm and
hospitality, which made this perfectly organised meeting a great success. Also, very
special thanks to Katharina Scherf for her invaluable help in proofreading. Finally, I
would like to express my appreciation to all friends, colleagues and sponsors for their
ongoing support of the PWG and the meeting.
Freising, March, 2017 Peter Koehler
1. Executive Summary
Among the topics of the meeting were food technological aspects of the production of
gluten-free baked goods, the importance of the small intestinal microbiome in the diet
of coeliac disease patients, analytical issues of gluten, clinical studies on coeliac
disease and non-celiac gluten sensitivity, serology of coeliac disease, further aspects of
the pathomechanism of coeliac disease, as well as legal issues.
Analytical session
Six presentations were given in this session. A novel reference material for barley
gluten based on C-hordeins was suggested. It appears that the immunodominant 33-
mer peptide is common among wheat cultivars but without being correlated with the
gluten content. Data on problems in the quantitation of the gluten content of wheat
starches induced lively discussions, which are also related to the future evaluation of
ELISA methods for approval with the Codex Alimentarius. This led to a meeting of
experts after the end of the conference to discuss about further actions. Finally,
breeding activities for wheat without coeliac disease activity and a possible role of
non-gluten proteins in coeliac disease were on the agenda.
Clinical session
This session included twelve presentations, which was by far the highest number
during the last years. Topics were widespread and included in vivo studies with
different diets in coeliac disease and non-celiac gluten sensitivity. Serological studies
showed that blood tests are now of major importance in the diagnosis of coeliac
disease. The issue of partially hydrolysed gluten for the immune system was
highlighted as well as the impact of amylase-trypsin inhibitors on intestinal
inflammations. Crystallographic studies on the interaction between HLA-DQ-gluten
and gluten-specific T-cell receptors gave insights into the pathomechanism of coeliac
disease.
Symposium: Enzymatic gluten degradation
The symposium comprised two presentations on the identification and use of enzymes
for degrading gluten and gluten peptides. A very interesting talk described the
identification of gluten-specific peptidases of the subtilisin family from dental plaque.
Some of these enzymes have a food-grade status and are promising candidates for
preparations that could be used for gluten ‘detoxification’ of foods or as oral
supplements for gluten degradation in the stomach. The second presentation dealt with
the application of peptide libraries to determine the stability of gluten peptides towards
gastrointestinal peptidases. Rat enzymes have been used so far, but the approach is
promising for the human peptidase system.
4. Analytical research reports
Alternatives for developing gluten-free bakery foods
Cristina M. Rosell
Institute of Agrochemistry and Food Technology (IATA-CSIC), Valencia, Spain
Introduction
Cereals, and more precisely wheat, have been at the base of the food pyramid through
the human history. Even today, cereals are the main players feeding human population;
although their worldwide contribution to nutrient uptake is different. However, there
are specific targeted groups with special requirements when consuming cereals,
namely gluten-containing grains. Coeliac disease, first considered to be a
gastrointestinal disease, is a gluten-sensitive enteropathy with genetic, immunologic,
and environmental bases. Great efforts are being made to understand the gluten-related
pathologies from the genetic and immunologic point of view and also the implication
of diet and gluten-free products on the life quality of the patients [1]. The clearest
statement is that the only way to ameliorate the symptoms is keeping a lifelong diet
free of gluten products.
In the last decade, gluten-free foods have shifted exponentially from a niche market to
become a revolution and to mark a lifestyle. Gluten-free has been described by
consumers as: “a mainstream sensation, embraced by both out of necessity and as a
personal choice toward achieving a healthier way to live”. However, in this scenario
nutritionists must play a fundamental role conducting counselling and closely
following the dietary management of coeliac individuals.
The initial challenge when developing gluten-free products as a necessity for solving
pathologies was to overcome the technological restrictions that the absence of gluten
provoked in the development of fermented cereal-based foods [2]. The main goal was
to look for tools to technologically replace the gluten giving sensorially accepted
products. However, gluten is not just a great protein matrix, it is a protein with
incomparable viscoelastic properties. Because of that its replacement has been an
enormous challenge during decades, and it is still a hot topic. Initially, only starches
and hydrocolloids were considered but later on, different tools have been developed
for defining food recipes resembling the quality of gluten-containing goods.
Nevertheless, in this picture not only the sensorial quality must be considered, it is an
essential requirement that those gluten-free foods provide the required nutrients’ intake
for those gluten-intolerants, contributing also to their wellbeing and healthy status at
present and also considering long-term nutrition.
Technological approaches for miming gluten in gluten-free bakery products
Replacement of gluten functionality has been a challenge for food technologists. Its
absence leads to less cohesive and elastic doughs that result in bread with a crumbling
texture, poor colour and low specific volume. Therefore, in the last years numerous
studies have been focused on improving the physical properties of gluten-free foods,
particularly fermented and baked foods like bread [3]. Gluten-free recipes are very
complex, and gluten-free bread is the result of the interaction of the ingredients.
Generally, bread development without gluten has involved the use of diverse
ingredients and additives with the purpose to obtain wheat bread-like properties.
Approaches proposed for obtaining gluten-free bread include the use of different
naturally gluten-free flours (rice, maize, sorghum, soy, buckwheat) and starches
(maize, potato, cassava, rice), dairy ingredients (caseinate, skim milk powder, dry
milk, whey), gums and hydrocolloids (guar and xanthan gums, alginate, carrageenan,
hydroxypropyl methylcellulose, carboxymethyl cellulose), emulsifiers (DATEM, SSL,
lecithins), non-gluten proteins from milk, eggs, legumes and pulses, enzymes
(cyclodextrin glycosyltranferases, transglutaminase, proteases, glucose oxidase,
laccase), and non-starch polysaccharides (inulin, galactooligosaccharides).
Strengthening additives or processing aids have been fundamental for miming gluten
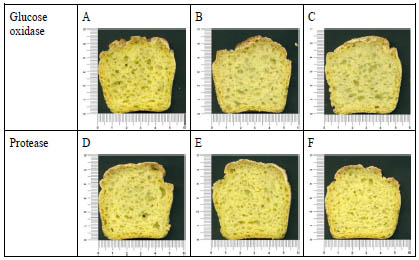
Figure 1. Cross section of corn breads obtained with different enzymes (glucose
oxidase and protease) at different levels (expressed as % (w/w) flour basis). Basic
recipe contained 1% xanthan gum. A: Glucose oxidase-0%, B: Glucose oxidase-
0.01%, C: Glucose oxidase-0.02%, D: Protease-0.05%, E: Protease-0.1%, F:
Protease-0.2%
viscoelastic properties [4]. With that purpose, mainly hydrocolloids have been used for
building an internal network able to hold the structure of fermented products. With the
same purpose, different crosslinking enzymes such as glucose oxidase,
transglutaminase and laccase have been used for obtaining a protein network within
the flour proteins [5]. Nevertheless, even the disruption of the flour proteins with
proteases has been revealed as a good strategy to improve dough performance and in
consequence the features of the resulting breads, owing to the decrease of protein
hydrophobicity [5]. However, it must be stressed that the effect of the enzymes as
gluten-free processing aids is greatly dependent on the type of flour, enzyme source
and level, which could lead to improve the bread performance or even to the opposite
effect (Fig. 1). Very often the combinations of ingredients and the optimization of the
breadmaking process can overcome the technological problems, yielding gluten-free
products that meet the consumer’s expectations concerning texture and appearance of
the fresh bread [6,7].
Nutritional and health aspects of gluten-free products.
Previous reviews showed that much research has been conducted on gluten-free foods
from different angles to obtain good quality gluten-free-foods. Nevertheless, the
nutritional quality of those products has been of interest only recently. In the last five
years, the driving force behind gluten-free research has been the nutritional quality.
Very recently, Matos and Rosell [8] reviewed the different available strategies for
improving the nutritional quality of gluten-free breads. The absence of gluten in
natural and processed foods constitutes the therapy treatment for coeliac disease,
which may lead to nutritional consequences linked to the composition of gluten-free
products. The exclusion of gluten-containing cereals, important vitamin and mineral
sources, from the diet might provoke deficiencies in iron, vitamin B and dietary fibre.
In fact, common nutrient deficiencies in coeliac subjects at diagnosis are
calory/protein, fibre, iron, calcium, magnesium, vitamin D, zinc, folate, niacin, vitamin
B12 and riboflavin [9]. Following a lifelong gluten-free diet requires a parallel
nutrition counselling, not only focused on the foods to avoid when sticking to a glutenfree
diet, but also the nutritional quality of gluten-free products must enter into the
equation to elude deficits and imbalances.
Some concerns have arisen after publishing some reports showing that the nutritional
quality of gluten-free products available on the market were poorer than their glutencontaining
counterparts. Gluten-free breads are starch based foods low in proteins and
high in fat content, with high glycaemic index [10]. Therefore, a lifelong adherence to
gluten-free products could lead to undernourishment and also mineral deficiencies that
might end in anaemia, osteopenia or osteoporosis. In the particular case of gluten
intolerance, it must also be considered of that coeliac disease induces an intestinal
lesion that leads to various deficiencies of nutrients, vitamins, and dietary minerals,
with ferropenia, vitamin B12, folic acid, and fat-soluble vitamin deficiencies being
especially frequent.
Therefore, a careful design of gluten-free bakery goods is needed for obtaining glutenfree
baked products resembling the nutritional composition of their gluten counterparts
to meet dietary guidelines without changing their dietary pattern and to avoid nutrient
deficiencies.
Enrichment or fortification is a strategy commonly applied to mitigate nutritional
deficiencies of the population and wheat flour has been a common carrier for minerals
and vitamins. In the case of gluten-free products, although this strategy has been less
exploited, there are some trends to complement or balance the nutritional composition
of those foods. In the case of minerals calcium salts like lactate, citrate, chloride and
carbonate have been proposed as sources of elementary calcium for obtaining fortified
gluten-free breads [11]. The supplementation of gluten-free bread with proteins has
been a technological strategy for improving the protein network and also for increasing
the nutritional quality of gluten-free breads. Legume flours have become very useful
for protein and fibre enrichment of bakery foodstuff, like gluten-free cakes, although it
is necessary to carefully select the legume to avoid any effect on the technological and
sensorial quality [12].
Lately, the physical treatment of the raw materials for enhancing the nutritional quality
or healthy pattern is gaining popularity. The selection of the particle size distribution
in the gluten-free flours has great impact on the technological properties of the
products, but it also determines the glycaemic index of the resulting fresh products.
For instance, in rice flour, particle size heterogeneity is responsible or different
patterns in starch enzymatic hydrolysis, allowing the modulation of their digestibility.
Particularly, enzymatic digestibility increases with the reduction of the particle size
[13]. With the same purpose, germination, toasting or cooking of the grains have been
proposed for increasing the nutritional, functional, and sensory properties of grains
such as pulses and cereals [14-15]. For instance germination of rice kernels under
controlled conditions of temperature and time allows the degradation of beta-glucans,
increases the content of certain essential amino acids and B-group vitamins and
improves protein and starch digestibility.
Further research
Currently, research is moving fast and numerous gluten-free foods are launched
annually. In spite of scientific advances, there is no date in the near future for having
high quality gluten-free food products nutritionally equivalent to gluten-containing
products. Lately, consumers’ interest in the role of nutrition for health and wellbeing
seems a priority. Therefore, today, the main concern of the industry is to innovate,
meet and satisfy consumer requirements. In the baking industry that trend has
prompted the development of baked goods keeping in mind the healthy concept.
Enrichment of formulations, physical treatment of raw materials and the usage of noncommon
flour sources are alternatives for enhancing the health benefits of gluten-free
baked foods. In that scenario, some other approaches like the exploration of the use of enzymes as “healthy aids” or the use of “smart starch” as vehicle of functional
ingredients must be encouraged [16-17].
Acknowledgements
The financial support of the Spanish Ministry of Economy and Competitiveness
(Project AGL2014-52928-C2-1-R) and the European Regional Development Fund
(FEDER) is acknowledged.
References
1. Arranz E, Fernandez-Bañares F, Rosell CM, Rodrigo L, Peña AS (eds): Advances
in the understanding of gluten related pathology and the evolution of gluten-free
foods. OmniaScience, Barcelona, Spain, 2015; Open access. http://www.
omniascience.com/monographs/index.php/monograficos/issue /view/24
2. Rosell CM, Barro F, Sousa C, et al. Cereals for developing gluten-free products
and analytical tools for gluten detection. J Cereal Sci 2014; 59: 354-364.
3. Houben A, Höchstötter A, Becker T. Possibilities to increase the quality in glutenfree
bread production: an overview. Eur Food Res Technol 2012; 235: 195-208.
4. Zannini E, Jones JM, Renzetti S, Arendt EK. Functional replacements for gluten.
Annu Rev Food Sci Technol 2012; 3: 227-245.
5. Renzetti S, Rosell CM. Role of enzymes in improving the functionality of proteins
in non-wheat dough systems. J Cereal Sci 2016; 67: 35-45.
6. Matos ME, Rosell CM. Quality indicators of rice based gluten free bread like
products: relationships between dough rheology and quality characteristics. Food
Bioprocess Technol 2013; 6: 2331-2341.
7. Matos ME, Rosell CM. Relationship between instrumental parameters and sensory
characteristics in gluten-free breads. Eur Food Res Technol 2012; 235: 107-489.
8. Matos ME, Rosell CM. A review: understanding gluten free dough for reaching
breads with physical quality and nutritional balance. J Sci Food Agric 2015; 95:
653-661.
9. Saturni L, Ferretti G, Bacchetti T. The gluten-free diet: Safety and nutritional
quality. Nutrients 2010; 2:16-34.
10. Matos ME, Rosell CM. Chemical composition and starch digestibility of different
gluten free breads. Plant Food Human Nutr 2011; 66: 224-230.
11. Krupa-Kozak U, Bączek N, Rosell CM. Application of dairy products as
technological and nutritional improvers of calcium-supplemented gluten-free
bread. Nutrients 2013; 5: 4503-4520. Open access.
12. Gularte MA, Gómez M, Rosell CM. Impact of legume flours on quality and in
vitro digestibility of starch and protein from gluten-free cakes. Food Bioprocess
Technol 2012; 5: 3142-3150.
13. de la Hera E, Rosell CM, Gómez M. Effect of water content and flour particle size
on gluten-free bread quality and digestibility. Food Chem 2014; 151: 526-531.
14. Cornejo F, Caceres PJ, Martínez-Villaluenga C, et al. Effects of germination on
the nutritive value and bioactive compounds of brown rice breads. Food Chem
2015; 173: 298-304.
15. Ouazib M, Garzón R, Zaidi F, et al. Germinated, toasted and cooked chickpea as
ingredients for breadmaking. J Food Sci Technol 2016; 53: 2664-2672.
16. Benavent-Gil Y, Rosell CM. Comparison of porous starches obtained from
different enzyme types and levels. Carbohydrate Polymers 2017; 157: 533-540.
17. Dura A, Yokoyama W, Rosell CM. Glycemic response to corn starch modified
with cyclodextrin glycosyltransferase and its relationship to physical properties.
Plant Foods Human Nutr 2016; 71: 252-258.
Detection of gluten in products containing barley: A
proposal for C-hordein as reference material
Xin Huang1, Päivi Kanerva2, Hannu Salovaara1, Tuula Sontag-Strohm1
1 Department of Food and Environmental Sciences, University of Helsinki, Helsinki,
Finland
2 Fazer Mills, Oy Karl Fazer Ab, Lahti, Finland
Introduction
When measuring residual barley prolamin (hordein) contamination in gluten-free
products by the R5 ELISA method, the concentration of prolamin is overestimated
with the gliadin standard [1-3]. The reason for this may be that the composition of the
gliadin standard is different from the composition of hordeins. A hordein standard is
needed for barley prolamin quantification instead of the gliadin standard. C-hordein,
the primary structure of which is almost entire repeats of PQQPFPQQ, is strongly
recognised by the R5 antibody and has 15-20 times more reactivity than the reference
gliadin [4]. The aim of this study was to investigate the proportion of C-hordein in
whole barley hordein, in order to explain the hordein overestimation with a gliadin
reference material in R5 antibody-based ELISA. An additional aim was to determine
whether a reference material using C-hordein could be used to quantify hordein, for
example, to determine the barley contamination in gluten-free ingredients and
products.
Materials and methods
Twenty-nine barley cultivars from Finland for feed and malt purposes were selected
for this study (Boreal Plant Breeding Ltd.). The total hordein of these cultivars were
extracted by 40% (v/v) aqueous 1-propanol with 5% (v/v) 2-mercaptoethanol, and the
hordein composition was determined by reversed-phase-HPLC by the peak area on a
C8 column. C-hordein, B-hordein and D-hordein were collected from the C8 column
and their protein content was determined with a bovine serum albumin standard.
Hordein fractions were analysed in a sandwich gliadin kit (R7006, R-Biopharm,
Darmstadt, Germany) to evaluate their immunoreactivities against the R5 antibody.
Barley flour cultivar Elmeri, Einar and Marthe with different C-hordein proportions
(33.1%, 25.6% and 17.4%) were selected for spiking in gluten-free oat flour (Provena,
Raisio Nutrition Ltd. Finland) to mimic the barley contamination in oat products. The
hordein concentration was determined by HPLC, R5 sandwich ELISA with gliadin
standard calibration, and with 40% C-hordein standard. The C-hordein was isolated
and purified in a preparative ion-exchange column and lyophilised, and 40% C hordein standard was prepared by mixing the protein solution of same concentration
4 : 6 (C-hordein : bovine serum albumin, which does not react with R5 antibody).
Results and discussion
The C-hordein content of whole hordein of the 29 cultivars ranged 2-fold, from 16.5%
to 33.1%. There was slight variation in C-hordein content of the same cultivar Elmeri
from 2010, 2014 and 2015 (33.1%, 29.2% and 28.1%). Taken the popularity of the
barley cultivars into account, the average C-hordein content of whole hordein in
Finland 2012-2015 was 25-26%. The corresponding protein group to the C-hordeins in
wheat are the ω1,2-gliadins, which shows about 70% sequence homology, with a
similar repetitive sequence in the central domain of PFPQQPQQ. The ω-gliadin
content of total gliadin has been reported to range from 6% to 20% [5], and from 10%
to 19% [6], which is in general lower than the content of C-hordein. The gliadin
standard contains 11.3% -gliadin of total gliadin by HPLC analysis [7].
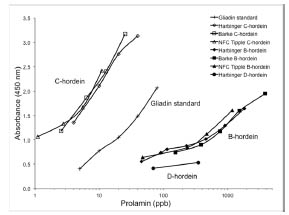
Figure 1. Reaction of isolated hordeins against R5 antibody in sandwich ELISA.
Three types of C-hordein and B-hordein were from cultivars Harbinger, Barke and
NFC Tipple. D-hordein was from cv. Harbinger
The reactivity of D-, C- and B-hordeins against the R5 antibody varied widely in
sandwich ELISA (Fig. 1). C-hordein was 10-20 times more reactive than the gliadin
standard, which in turn was 8-25 times more reactive than B-hordein. The slope of the
curve indicated that C-hordein and gliadin standard had similar affinity with the R5
antibody, while B-hordein had less, and D-hordein had almost none. The three types of
C-hordein reacted similarly with R5 antibody, although their HPLC patterns were
different, as well as three types of B-hordein. The varying reactivity of hordein
subunits against the R5 antibody is attributable to the number of epitopes. The main
R5 epitope, QQPFP, appeared 13 times in C-hordein (Uniprot Q40055), and minor
epitopes QQPYP, QQTFP, PQPFP and QLPFP appeared once each. One main QQPFP
epitope and 7 minor epitopes were found in B3 hordein (Uniprot I6TEV5), and 5
QQPFP epitopes in B1 hordein (Uniprot P06470). Only one QQPFP epitope was
found in γ3-hordein (Uniprot P80198) and no R5 epitope was found in D-hordein
(Uniprot Q84LE9) [8].
In sandwich ELISA, the affinity (the slope) of C-hordein with R5 antibody behaved
similarly to the gliadin standard, and at a ratio of 3 C-hordein : 7 bovine serum
albumin (30% C-hordein), the reaction almost matched that of the gliadin standard
(Fig. 2). The curves of purified whole hordein of common cultivars, such as cv. Barke
and NFC tipple (C-hordein proportions 24.5% and 28.1% respectively), were above
that of the gliadin standard and between that of the 30% and 50% C-hordein standard.
The curve of cv. KWS Asta, with its low C-hordein proportion (16.5%), was close to
the gliadin standard curve and that of 30% C-hordein. The whole hordein of a barley
cultivar with low C-hordein content acted like wheat gliadin against R5 antibody,
however, the barley cultivars usually have higher C-hordein content than that. Chordein
mixed with inert protein in the right ratio presented the whole hordein in R5
analysis.

Figure 2. Reaction of purified whole hordein of 6 cultivars in R5 sandwich ELISA
compared with 30%, 40% and 50% C-hordein standards and gliadin standard
When measuring the prolamin concentration of prolamin in barley-contaminated oats,
with the 40% C-hordein standard, the estimated prolamin concentration was 1.2 times
(cv. Elmeri), 0.85 times (cv. Einar) and 0.63 times (cv. Marthe) the HPLC results,
however, the concentration calibrated by gliadin standard was 2.5 times (cv. Elmeri),
1.8 times (cv. Einar), and 1.2 times (cv. Marthe) the HPLC results (Fig. 3). For cv.
Elmeri and Einar, the estimated value by the 40% C-hordein standard were not
significantly different from those determined by HPLC, but for cv. Marthe the estimate
was significantly lower, until the standard was changed to 30% C-hordein.
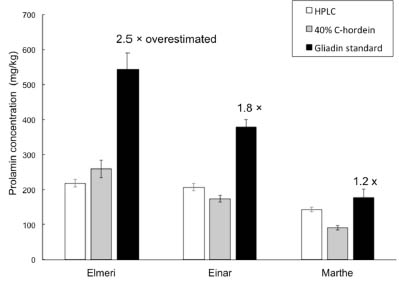
Figure 3. Prolamin concentration of gluten-free oat flour spiked with three barley
flours, determined by HPLC, R5 sandwich ELISA with 40% C-hordein standard, and
R5 sandwich ELISA with gliadin standard. Error bars show standard error
Conclusion
This study determined that the high proportion of C-hordein in total hordein is the
reason for the consistent overestimation of hordein by the R5 ELISA assay which uses
gliadin as reference material in gluten-free analysis. We isolated C-hordein and
propose it as the reference material for quantifying hordein concentration in glutenfree
food originated from barley, including those that may have been contaminated
with barley.
References
1. Kanerva PM, Sontag-Strohm TS, Ryöppy PH, et al. Analysis of barley
contamination in oats using R5 and ω-gliadin antibodies. J Cereal Sci 2006; 44:
347-352.
2. Hernando A, Mujico JR, Mena MC, et al. Measurement of wheat gluten and
barley hordeins in contaminated oats from Europe, the United States and Canada
by Sandwich R5 ELISA. Eur J Gastroenterol Hepatol 2008; 20; 545-554.
3. Mujico JR, Mena MC, Lombardía M, et al. On the way to reliable quantification
of barley hordeins using the R5 ELISA technique. In: Stern M (eds): Proceedings
of the 22nd meeting working group on prolamin analysis and toxicity. Verlag
Wissenschaftliche Scripten, Zwickau, Germany, 2008; pp. 29-34.
4. Huang X, Kanerva PM, Salovaara HO, et al. Degradation of C-hordein by metalcatalysed
oxidation. Food Chem 2016; 196: 1256-1263.
5. Wieser H, Seilmeier W, Belitz HD. Quantitative determination of gliadin
subgroups from different wheat cultivars. J Cereal Sci 1994; 19: 149-155.
6. Daniel C, Triboi E. Effects of temperature and nitrogen nutrition on the grain
composition of winter wheat: effects on gliadin content and composition. J Cereal
Sci 2000; 32: 45-56.
7. Van Eckert R, Berghofer E, Ciclitira PJ, et al. Towards a new gliadin reference
material-isolation and characterisation. J Cereal 2006; 43: 331-341.
8. Tanner GJ, Blundell MJ, Colgrave, ML, et al. Quantification of hordeins by
ELISA: The correct standard makes a magnitude of difference. PLoS One 2013;
8(2): e56456.
Quantitation of the 33-mer peptide from α-gliadins in
wheat flours by LC-MS/MS
Kathrin Schalk, Christina Lang, Herbert Wieser, Peter Koehler, Katharina A. Scherf
Deutsche Forschungsanstalt für Lebensmittelchemie, Leibniz Institut, Freising,
Germany
Introduction
The dietary intake of storage proteins (gluten) from wheat (gliadins, glutenins), rye
(secalins), and barley (hordeins) is known to cause coeliac disease (CD) in genetically
predisposed individuals. A strict lifelong gluten-free diet is currently the only available
therapy. All gluten proteins contain CD-active epitopes [1], which are resistant to
cleavage by human gastric, pancreatic, and brushborder enzymes. A 33-mer peptide
from α2-gliadin (LQLQPFPQPQLPYPQPQLPYPQPQLPYPQPQPF) was shown to
survive gastrointestinal digestion and has frequently been described as most
immunodominant gluten peptide [2,3], because it comprises three overlapping
DQ2.5/T-cell epitopes, PFPQPQLPY (DQ2.5-glia-α1a, one copy), PYPQPQLPY
(DQ2.5-glia-α1b, two copies), and PQPQLPYPQ (DQ2.5-glia-α2, three copies) [1].
Due to its unique structure, the 33-mer peptide plays an important role in the scientific
literature with 570 results for a search in the database ScienceDirect with “33 mer” and “coeliac disease” as keywords (as of October 29, 2016). The 33-mer was also used as
an antigen to produce two monoclonal antibodies (A1 and G12), which are now used
in enzyme-linked immunosorbent assays to determine gluten contents in foods labelled
as gluten-free [4].
DNA-sequencing of eleven α-gliadins (α1 - α11) from the Norwegian common (bread)
wheat (Triticum aestivum) cultivar (cv.) Mjølner (MJO) revealed that only α2-gliadin
contained the 33-mer amino acid sequence at positions 56 - 88 [5]. According to a
BLAST search in the UniProtKB database within 587 entries for α-gliadins from
Triticum sp., the 33-mer sequence (100% identity) was found in only 17 protein
sequences from T. aestivum and in three from T. spelta (as of September 13, 2016). Of
these 20 sequences, only three have evidence at transcript level (Q9M4L6, Q1WA39
and A5JSA6) inferred from three Chinese wheat cultivars, but only one (P18573) has
evidence at protein level based on data of the Norwegian wheat cv. MJO. Despite the
high number of papers featuring the 33-mer, there is no information on the presence
and quantities of the 33-mer peptide in different wheat species and cultivars.
Therefore, the aim of the present study was to develop a stable isotope dilution assay
(SIDA) combined with liquid chromatography tandem mass spectrometry (LCMS/
MS) for the determination of the presence and quantity of the 33-mer. Fifty-seven
flours of different wheat species from around the world were investigated, including
hexaploid common wheat (T. aestivum) and spelt (T. aestivum ssp. spelta), tetraploid durum wheat (T. turgidum durum) and emmer (T. turgidum dicoccum), and diploid
einkorn (T. monococcum) to assess the importance of this CD-active peptide.
Materials and Methods
Preparation and characterization of flour samples
Twenty-three modern and 15 old (year of first registration before 1950) common
wheat cultivars from different harvest years grown worldwide, and one rye cultivar
(cv. Visello, harvested in 2013) were either obtained as flours or milled on a
Quadrumat Junior mill (Brabender, Duisburg, Germany) and sieved (mesh size 0.2
mm). Two spelt, durum wheat, emmer, and einkorn cultivars each were milled on a
Laboratory 3100 cross beater mill (Perten Instruments, Hamburg, Germany) to
wholemeal flours.
The crude protein content (nitrogen content x 5.7) of the flours was determined by the
Dumas combustion method. The contents of albumins/globulins, α-gliadins, gliadins,
glutenins, and gluten (sum of gliadins and glutenins) were determined by modified
Osborne fractionation of the flours followed by RP-HPLC-UV (210 nm) analysis [6].
Sample preparation
The flours (150 - 200 mg) were defatted with pentane/ethanol (95/5, v/v; 2 x 2.0 mL).
After removal of the albumins/globulins, the gliadins were extracted with 60% (v/v)
ethanol, dried, and re-suspended in a TRIS-HCl-buffer (pH 7.8). The stable isotope
labelled standard (*33-mer, LQLQP*FPQPQLPYPQPQLPYPQPQLPYPQ*PQ*P*F,
with *F: L-[13C9][15N]-phenylalanine and *P: L-[13C5][15N]-proline) was added (3 μg)
and the gliadin-peptide mixture hydrolysed with α-chymotrypsin (enzyme-to-protein
ratio of 1:200) for 24 h at 37 °C. Trifluoroacetic acid (5 μL) was added to stop the
digestion. The peptide mixture was dried, re-dissolved in formic acid (FA) (0.1%, v/v,
500 μL), filtered (0.45 μm) and analysed by LC-MS/MS.
LC-MS/MS
A triple-stage quadrupole mass spectrometer (TSQ Vantage, Thermo Fisher Scientific,
Dreieich, Germany) was used in the ESI positive mode. The mass spectrometer was
operated in the multiple reaction monitoring (MRM) mode using the most abundant
MRM transition as quantifier and the three MRM transitions following in abundance
as qualifiers (Tab. 1). A declustering voltage of -10 V was set for all transitions. The
33-mer and the labelled *33-mer peptides were dissolved in FA (0.1%, v/v,
10 μg/mL). These two stock solutions were mixed in molar ratios n (*33-mer)/n (33-
mer) between 9.2 and 0.02 (1+9, 1+4, 1+3, 1+1, 3+1, 4+1, 9+1, 14+1, 19+1, 29+1, and
39+1) for calibration. An UltiMate 3000 HPLC system (Dionex, Idstein, Germany)
was coupled to the mass spectrometer equipped with an XBridge Peptide 3.5 μm BEHC18
column (1.0 x 150 mm, 13 nm; Waters, Eschborn, Germany). The LC conditions
were set as follows: solvent A, FA (0.1%, v/v) in water , solvent B, FA (0.1%, v/v) in acetonitrile; gradient 0 - 5 min 5% B, 5 - 22 min 5 - 55% B, 25 - 30 min 90% B; 30 -
35 min 90 - 5% B, 35 - 45 min 5% B, flow rate, 0.1 mL/min; injection volume, 10 μL,
column temperature, 22 °C.
Table 1. Multiple reaction monitoring (MRM) parameters of the 33-mer peptide and
the stable isotope labelled *33-mer peptide.
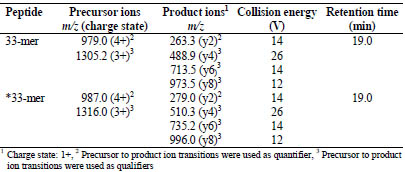
The limits of detection (LOD) and quantitation (LOQ) of the LC-MS/MS method for
the 33-mer peptide were determined. Rye flour was used as blank, because of the
absence of α-gliadins. The rye prolamin extract was spiked at 7 different concentrations
(0.1 - 200 mg/kg) of 33-mer peptide and the samples were prepared and analysed
as described above. The LOD was calculated based on a signal-to-noise ratio (S/N) of
3, and the LOQ on an S/N ratio of 10.
Statistics
Linear Pearson’s product moment correlations were calculated between contents of 33-
mer and α-gliadins, gliadins, gluten or crude protein for all analysed wheat and spelt
cultivars. Principal component analysis (PCA) was carried out with XLStat 2016
(Addinsoft, New York, NY, USA) to determine if the contents of 33-mer, α-gliadins,
gliadins, gluten, and crude protein could be used to differentiate between spelt, modern
and old common wheat cultivars.
Results and Discussion
A [13C28]- and [15N4]-labelled *33-mer peptide (LQLQP*FPQPQLPYPQPQLPYPQ
PQLPYPQ*PQ*P*F, with *F: L-[13C9][15N]-phenylalanine and *P: L-[13C5][15N]-
proline, monoisotopic mass 3943.0) was used as stable isotope labelled internal
standard. It differed in 32 mass units compared to the unlabelled analyte (33-mer,
monoisotopic mass 3911.0). Based on the fragmentation pattern of the 33-mer, the
[13C]/[15N]-labelled amino acids were positioned in such a way that the label remained
in the product ions (Tab. 1). The response factor was determined using the peak area
ratio A (*33-mer)/A (33-mer) at different values of n (*33-mer)/n (33-mer) between 0.02 and 9.2 within the linear range based on the MRM transitions m/z 987.0 279.2
(*33-mer) and m/z 979.0 263.3 (33-mer). As expected from SIDA, the response
factor was 0.999. The LOD of the LC-MS/MS method to detect the 33-mer peptide
was 13.1 μg/g rye flour and the LOQ was 47.0 μg/g rye flour.
The 33-mer was determined in flours of 23 modern and 15 old common wheats from
different harvest years and two spelt cultivars. In this context, old common wheat is
defined as a cultivar from T. aestivum with its year of first registration prior to 1950.
All flours were characterised including determination of crude protein contents and
quantitation of α-gliadins, gliadins, glutenins, and gluten after modified Osborne
fractionation combined with RP-HPLC [6,7].
The 33-mer was present in all common wheat and spelt flours in a range from 90.9 to
602.6 μg/g of flour (Fig. 1A). Overall, the modern wheat cv. Yumai-34 (harvested in
2014, Y14) had the highest (602.6 μg/g flour) and the old wheat cv. Ackermanns
Brauner Dickkopf (ABD) the lowest (90.9 μg/g flour) amount of 33-mer. Most of the
modern and old wheat flours contained the 33-mer in a range of 200 - 400 μg/g flour
with an overall average of 368 ± 109 μg/g flour. Special attention was directed to cv.
MJO, because the 33-mer was first identified in this cultivar [5], which had a 33-mer
content of 515.0 μg/g flour. A certain trend, e.g., that modern wheat cultivars generally
contain higher amounts of 33-mer than old wheat or spelt cultivars could not be
derived from the data.
The 33-mer contents of all analysed flours were also calculated based on the amount of α-gliadins (Fig. 1B). MJO had the highest content of 33-mer in α-gliadins (23.2 mg/g α-gliadins) caused by the high 33-mer content and the low amount of α-gliadins
(2.2%) in flour. ABD had the lowest amount of 33-mer in α-gliadins (4.1 mg/g α-
gliadins). The overall average content was 11.7 ± 3.1 mg/g α-gliadins. Because there is
virtually no data in the literature, it was difficult to compare these values with earlier
studies, but one paper by van den Broeck et al. on the quantitation of the 33-mer using
LC-MS with external calibration found comparable values for two wheat cultivars [8].
Correlations and PCA
The 33-mer contents of the 51 modern and old common wheat and spelt cultivars
(based on flour) were correlated to the contents of α-gliadins, gliadins, gluten, and
crude protein. A weak correlation (r = 0.568) was observed between 33-mer and α-
gliadin contents, but there was no correlation to gliadin contents (r = 0.469), gluten
contents (r = 0.526) or crude protein contents (r = 0.466).
PCA with 33-mer, α-gliadins, gliadins, gluten, and crude protein contents of the 51
flours was performed to assess whether these variables could be used to differentiate
between spelt, modern common wheat, and old common wheat cultivars (Fig. 2).
However, PCA revealed that these five variables were unsuitable to differentiate
between spelt, modern common wheat, and old common wheat cultivars. Five old
common wheat cultivars were placed on the far left, but the other ten old cultivars were located right in the middle at similar coordinates as the modern common wheat
cultivars. The two spelt cultivars were also situated in between the common wheat cultivars.
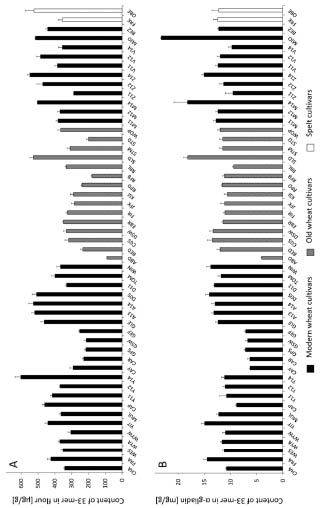
Figure 1. Contents of 33-mer based on flour [μg/g] (A) and based on α-gliadins
[mg/g] (B). 23 modern and 15 old common wheat cultivars (49 samples in total due to
multiple harvest years) and two spelt cultivars were analysed. Wheat cultivars
registered prior to 1950 were designated as old
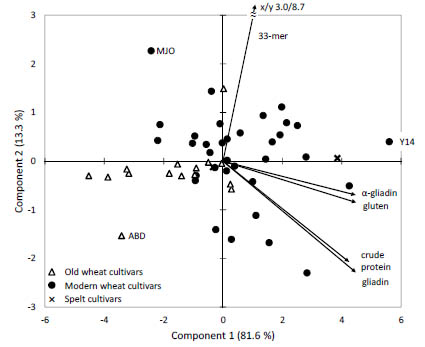
Figure 2. Principal component analysis biplot of data for 33-mer, α-gliadin, gliadin,
gluten, and crude protein contents. 23 modern and 15 old common wheat (49 samples
in total due to multiple harvest years) and two spelt cultivars were analysed. Wheat
cultivars registered prior to 1950 were designated as old. ABD, wheat cv. Ackermanns
Brauner Dickkopf, MJO, wheat cv. Mjølner, Y14, wheat cv. Yumai-34, harvest year
2014)
The 33-mer peptide was also analysed in two durum wheat and two emmer cultivars
(genome AABB) as well as in two diploid einkorn cultivars (genome AA), but it was
not detected in each of these wheat species (< LOD). In comparison to hexaploid
common wheat, durum wheat, emmer, and einkorn do not contain the D-genome,
which originated from hybridisation of T. turgidum dicoccum (genome AABB) with
Aegilops tauschii (genome DD). The absence of the 33-mer peptide can be explained
by the fact that this peptide is encoded by genes located in the Gli-2 locus on
chromosome 6D, which is missing in durum wheat, emmer, and einkorn [5].
Conclusion
This is the first study to establish a SIDA combined with LC-MS/MS to quantitate the
immunodominant 33-mer peptide from α2-gliadin in wheat flours. All 40 modern and
old common wheat and spelt cultivars analysed contained the 33-mer peptide (51 flour
samples in total, because several flours were available from different harvest years).
The special attention paid to this peptide in the scientific literature seems to be
legitimated not only because of its unique structure containing six copies of three
overlapping coeliac-active epitopes, but also because of its presence in all hexaploid
wheat cultivars analysed in this study. Further work will focus on correlating the 33-
mer content analysed by LC-MS/MS with the gluten content determined by ELISA
using the G12 antibody, which was raised against the 33-mer.
Acknowledgement
The authors would like to thank Andreas Börner (Leibniz Institute of Plant Genetics
and Crop Plant Research, Resources Genetics and Reproduction, Gatersleben,
Germany), Friedrich Longin (University of Hohenheim, LSA - Resarch Group Wheat,
Stuttgart, Germany), Anette Moldestad (Nofima, Ås, Norway), Roland Poms (Imprint
Analytics, Neutal, Austria), Sándor Tömösközi (Budapest University of Technology
and Economics, Department of Applied Biotechnology and Food Science, Budapest,
Hungary), and Bin Xiao Fu (Canadian Grain Commission, Grain Research Laboratory,
Winnipeg, Canada) for providing wheat grains and flours.
References
1. Sollid LM, Qiao S-W, Anderson RP, et al. Nomenclature and listing of celiac
disease relevant gluten T-cell epitopes restricted by HLA-DQ molecules.
Immunogenetic 2012; 64: 455-460.
2. Shan L, Molberg Ø, Parrot I, et al. Structural basis for gluten intolerance in celiac
sprue. Science 2002; 297: 2275-2279.
3. Shan L, Qiao S-W, Arentz-Hansen H, et al. Identification and analysis of
multivalent proteolytically resistant peptides from gluten: implications for celiac
sprue. J Proteome Res 2005; 4: 1732-1741.
4. Morón B, Cebolla A, Manyani H, et al. Sensitive detection of cereal fractions that
are toxic to celiac disease patients by using monoclonal antibodies to a main
immunogenic wheat peptide. Am J Clin Nutr 2008; 87: 405-414.
5. Arentz-Hansen H, McAdam SN, Molberg Ø, et al. Production of a panel of
recombinant gliadins for the characterisation of T cell reactivity in coeliac disease.
Gut 2000; 46: 46-51.
6. Wieser H, Antes S, Seilmeier W. Quantitative determination of gluten protein
types in wheat flour by reversed-phase high-performance liquid chromatography.
Cereal Chem 1998; 75: 644-650.
7. Schalk K, Lang C, Wieser H, et al. Quantitation of the immunodominant 33-mer
peptide from α-gliadin in wheat flours by liquid chromatography tandem mass
spectrometry. Sci Rep 2017; doi: 10.1038/Srep45092.
8. van den Broeck HC, Cordewener JHG, Nessen M, et al. Label free targeted
detection and quantification of celiac disease immunogenic epitopes by mass
spectrometry. J Chrom A 2015; 1391: 60-71.
The gluten content of wheat starches
Tanja Šuligoj, H. Julia Ellis, Paul J. Ciclitira
Department of Gastroenterology, Kings College, St Thomas Hospital, London, United
Kingdom
Introduction
The only generally accepted treatment for coeliac disease (CD) is a lifelong strict
gluten-free diet that involves avoidance of wheat, rye and barley. Wheat gluten
contains gliadin, low (LMWG) and high (HMWG) molecular weight glutenin proteins,
all three of which have been shown to be CD-toxic [1-3]. Many gluten-free foods are
available. This includes those that are commercially marketed, 80% of which in the
UK are based on purified wheat starch. Foods that are supplied as gluten-free are
required to contain <20 mg/kg gluten and those that are labelled "very low gluten" 21-
100 mg/kg gluten. The only FAO certified assay to quantify the gluten content of
foods for individuals with CD is based on the R5 monoclonal antibody (mAb) that
recognises gliadin but not glutenin [4]. The value for the gluten content of a given food
for this assay is determined by quantifying the gliadin content and multiplying the
value by two to yield the gluten content of a given food. This extrapolation, based on
the gliadin content may be invalid due to the differing solubility of gluten proteins,
that is gliadin and glutenins, when food is processed.
Aims
We wished to improve the extent and accuracy of quantification of CD-triggering
peptides in purified wheat starch that is a common ingredient of many commercially
available processed gluten-free foods for individuals with CD.
Materials and methods
We have generated three mAbs to wheat gluten proteins. This includes PN3 to wheat
gliadin that was raised against and detects coeliac-toxic A-gliadin AA31-49 [5,6],
CDC5 to the CD-toxic immunodominant epitope in wheat gliadin that was raised
against and detects α2-gliadin AA57-75 [7] and CDC7 to wheat glutenin generated to
the protein 1Dy10 HMWG glutenin subunits (HMWG) [8]. We developed three
separate competitive ELISAs employing the three separate mAbs, PN3, CDC5 and
CDC7. We assessed the gluten content of three wheat starches termed A, B & C that
are supplied as standards for the Transia kit that is marketed to quantify the gluten
content of foods based on the use of a mAb raised against ω-gliadin [9].
Results and discussion
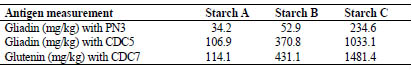
Gliadin contents of wheat starches A, B and C were 34.2, 52.9 and 234.6 mg/kg as
determined by PN3 mAb. Gliadin contents of the wheat starches measured using
CDC5 mAbs followed the same trend of increasing gliadin content from starch A to C,
but the values were higher. Gliadin contents were 106.9, 370.8 and 1033.1 mg/kg.
Starches A, B and C contained 114.1, 431.1 and 1481.4 mg/kg glutenin as assessed
with CDC7 mAbs (Tab. 1).
Table 1. Gliadin and glutenin content (in mg/kg) of wheat starches A, B and C as
determined with PN3, CDC5 and CDC7 mAb.
Gluten contents were then calculated based on measurements of the three mAbs. Two
approaches were undertaken: a) by following the standard method of extrapolating
gliadin content to total gluten by multiplying the gliadin content by factor 2 ; b) by
summing up gliadin and glutenin content to obtain the gluten content. Two different
results were obtained, depending on whether gliadin content was measured with PN3
or CDC5 mAb (Tab. 2).
When PN3 mAb measurement was used to extrapolate the gliadin content of starch A
to total gluten, the obtained value was 68.4 mg/kg gluten which is within the limit for “very low gluten” labelling of foodstuffs. When another anti-gliadin mAb (CDC5) was
used for the same starch, the gluten content was more than 3 times higher (213.8
mg/kg), exceeding the 100 mg/kg cut-off value for “very low gluten”.
Summing up the values of gliadin and glutenin measurements to obtain total gluten led
to two different results: 148.3 and 221 mg/kg depending on whether values of PN3 or
CDC5 measurements were taken to be summed up with CDC7 measurements (Tab. 2).
Interestingly, the calculation for total gluten based on the approach gluten = 2 x gliadin
(PN3) was more than 2-fold lower than when gluten was calculated by summing up
gliadin (PN3) plus glutenin (CDC7) which equalled 148.3 mg/kg for wheat starch A.
On the contrary, for CDC5 mAb these two approaches resulted in very similar final
gluten contents (213.8 and 221 mg/kg respectively) (Tab. 2).
Similarly, when the gliadin content of wheat starch B was extrapolated to total gluten
(by multiplying the gliadin content by 2), the obtained value was 105.8 mg/kg for PN3
measurement and 7-fold higher gluten content (741.6 mg/kg) was seen for CDC5
measurement. When total gluten of starch B was obtained by the other approach, i.e.
summing up the values of gliadin and glutenin (CDC7) measurements, they resulted in
484 and 801.9 mg/kg gluten for PN3 and CDC5 measurements, respectively. Gluten
content calculated by gluten = 2 x gliadin as opposed to gluten = gliadin + glutenin (CDC7) differed 4.6-fold for PN3 mAb measurements and 1.1-fold for CDC5, the
higher values obtained by the gluten = gliadin + glutenin approach (Tab. 2).
The results for wheat starch C had a similar trend. Gluten content obtained by
multiplying gliadin measurements by factor 2 resulted in 4.4-fold higher total gluten
content for CDC5 measurement (2066.2 mg/kg) than PN3 measurement (469.2
mg/kg). The other approach whereby glutenin content (obtained with CDC7
measurement) was summed up with gliadin content resulted in 3.6-fold increase of
gluten content for PN3 mAb measurements (from 469.2 to 1716 mg/kg) and 1.2-fold
increase of gluten content for CDC5 mAb measurements (from 2066.2 to 2514.5
mg/kg) (Tab. 2).
Table 2. Gluten content (in mg/kg) of wheat starches A, B and C as determined by
multiplying gliadin content by 2 versus summing up the measurements of gliadin and
glutenin content.

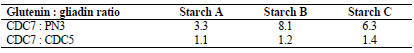
Further, ratios of glutenin to gliadin content of the wheat starches were determined by
dividing the glutenin values obtained with CDC7 mAb by gliadin values assessed
either by PN3 or CDC5 mAb (Tab. 3).
Table 3. Ratios of glutenin to gliadin contents in wheat starches A, B and C depending
on which gliadin monoclonal antibody (PN3 or CDC5) is used for comparison with
CDC7 mAb measurements.
Gliadin content of starches A, B and C depended on whether PN3 or CDC5 was
chosen for the measurement (Tab. 1). Ratios of gliadin contents in the three wheat
starches were determined by dividing the gliadin content obtained with CDC5 by
gliadin content obtained by PN3 mAb. The scale of difference in gliadin content varied
amongst starches (3.1- to 7.0-fold) (Tab. 4).
Table 4. Ratios of gliadin contents in wheat starches A, B and C as obtained with the
two gliadin antibodies (CDC5 and PN3).

Discussion
We demonstrated that a broadened repertoire of mAbs specific for CD-triggering
peptides enabled improved measurement of gluten in foods, by allowing a more
realistic measurement of the CD-triggering epitopes within the glutenins. This applied
particularly to the measurements where PN3 mAb was used for measuring gliadin
content and then obtaining total gluten either by the “standard method“ or by summing
up gliadin and glutenin measurements. This observation was less applicable to CDC5
measurements as multiplying gliadin content obtained by CDC5 mAb by a factor 2
differed very little to obtaining gluten content by summing up its gliadin content with
glutenin. The total gluten based on CDC5 and CDC7 mAb measurements indicate that
the epitopes that these two mAbs detect were more equally distributed as opposed to
those detected by PN3 and CDC7 mAbs in the three starches.
None of the three wheat starches were “gluten-free”, as they all contained more than
20 mg/kg gluten. When anti-gliadin mAb PN3 was used for the measurements of
gluten contamination of starch A it resulted in values that would classify it as “very
low gluten” foodstuff as it contained less than 100 mg/kg gluten. This was applicable
for the “standard” method of determining the total gluten by multiplying the gliadin
value by factor 2. However, when total gluten content was assessed by summing up
the values obtained with PN3 mAb and anti-glutenin mAb CDC7, the gluten values
were well above the cut-off value for “very low gluten”. This data clearly
demonstrates the importance of measuring both groups of proteins in gluten
responsible for CD toxicity (gliadins and glutenins). This is particularly important for
processed foodstuffs like wheat starches where gliadin : glutenin ratios have been
shown to vary greatly [10,11].
Glutenin to gliadin ratios of the wheat starches varied between 1.1-8.1. Our results
demonstrate that multiplying gliadin content by factor 2 to estimate for the glutenin
may be invalid for processed foodstuffs, which is in agreement of Wieser and
Koehler’s [10] observations. Measurement of gliadin alone therefore cannot predict
total gluten content in foods. The standard method of multiplying gliadin content by 2
would lead to gross underestimation of gluten content in our wheat starches if the
antibody used for detection of gliadins was PN3 mAb.
The lower glutenin to gliadin ratios were obtained when comparing the glutenin
contents with gliadin determined with CDC5 mAbs. The higher ratio of glutenin to
gliadin obtained with PN3 as anti-gliadin antibody can be explained by the lower
amounts of the gliadin peptide that PN3 detected in the wheat starches. This was
further confirmed by calculating the gliadin to gliadin ratios determined by PN3 and
CDC5 antibodies which showed that the amount of detected gliadin in a foodstuff
depends greatly on which antibody is used for quantification. This concept was
demonstrated previously in a study of van Eckert et al. [12] who showed that two
different anti-gliadin antibodies (PN3 and R5) reacted with different individual
proteins in different protein sub-fractions of the reference gliadin separated by two
dimensional electrophoresis.
Further, our results differed from the reference values for gluten contamination of the
three wheat starches A, B and C. The reference values were provided in the
manufacturer’s information sheet and had been obtained using a monoclonal antibody
which detects ω-gliadins and were as follows: <100 mg/kg gluten for starch A, 300 -
600 mg/kg gluten for starch B and 1000 - 2500 mg/kg for starch C [9]. The value of <100 mg/kg complied with the previous regulations for labelling foods to be glutenfree
but not with the current regulations of <20 mg/kg. Our results of gluten
contamination did follow the same trend of increasing gluten contamination from
starch A to C, but the values were not the same. Gluten contamination assessment with
antibodies of different specificity can therefore result in different gluten amounts,
which is consistent with Allred and Ritter’s observations [13]. Of note, the
manufacturer of the three starches A, B and C did not provide information as to which
wheat cultivars the starches were obtained from. In this respect we do not know
whether the flour from the same cultivar was used and resultant starches subjected
further to three different washing processes or whether three different cultivars were
used.
There is a dilemma in the field of gluten measurement of what should be quantified in
order to assess the overall toxicity of foods for CD sufferers [14]. There are several
CD-triggering epitopes [15]. It is probably unrealistic to detect all of them. The gliadin
fraction of wheat gluten has long been established as CD-triggering. However, the
glutenins have only recently been shown to exacerbate the disease. Our monoclonal
antibodies detect CD-triggering epitopes distributed amongst both groups of proteins,
for which there is substantial clinical data confirming their role in CD pathogenesis. It
is interesting, although not surprising, that contamination of wheat starches with
glutenins was notably higher than with gliadins. This is likely due to different
solubility characteristics of the gluten protein fractions as a result of food processing
[13,16,17]. The glutenins are less water-soluble and therefore more likely to stay
adsorbed to starch granules after washing [16]. It is therefore crucial to detect glutenin
contamination of processed foodstuffs and thereby improve the extent of measured
gluten components. Our findings are consistent with Allred [13] who demonstrated
that all processed foodstuffs (n = 40) tested in their study contained 4- to 10-fold
higher gluten values when assessed with the mAb that has high affinity to glutenins as
opposed to R5 mAb with high affinity for gliadins.
Conclusion
We suggest that a broadened repertoire of mAbs specific for CD-triggering peptides
enables improved measurement of gluten in foods for individuals with CD, by
allowing a more realistic measurement of the CD-triggering epitopes within the
glutenins. The total gluten content depended on the specificity of the mAb(s) used for
quantitation. In addition, the glutenin to gliadin ratios varied greatly between wheat
starches. We therefore suggest that multiplying the gliadin content by a factor of 2 toestimate the total gluten content of a given nominally gluten-free food, particularly
those that are based on purified wheat starch may be invalid for processed foodstuffs.
Acknowledgements
The authors wish to thank the Rosetrees Trust and Clinical Research Trust for support
References
1. Ciclitira PJ, Evans DJ, Fagg NL, et al. Clinical testing of gliadin fraction in
coeliac patients. Clin Sci 1984; 66: 357-364.
2. Vader W, Kooy Y, van Veelen P, et al. The gluten response in children with
coeliac disease is directed towards multiple gliadin and glutenin peptides.
Gastroenterol 2002; 122: 1729-1737.
3. Dewar DH, Amato M, Ellis HJ, et al. The toxicity of high molecular weight
glutenin subunits of wheat to patients with coeliac disease. Eur J Gastroenterol
2006; 18: 493-491.
4. Osman AA, Uhlig HH, Valdes I, et al. A monoclonal antibody that recognises a
potential coeliac-toxic repetitive pentapeptide epitope in gliadin. Eur J
Gastroenterol Hepatol 2001; 13:1189-1193.
5. Freedman AR, Galfre G, Gal E, et al. Monoclonal antibody ELISA to quantitate
wheat gliadin contamination of gluten-free foods. J Immunol Methods 1987; 98:
123-127.
6. Ellis HJ, Rosen-Bronson S, O‘Reilly R, et al. Measurement of gluten using a
monoclonal antibody to a coeliac toxic peptide of A-gliadin. Gut 1998; 43:190-
195.
7. Ellis HJ, Bermudo Redondo M, Šuligoj T, et al. Gluten quantification of foods via
the immunodominant gliadin epitope. F Nutr Rep 2016; 1(3): 1-7.
8. Ellis HJ, Bermudo Redondo C, Šuligoj T, et al. Monoclonal Antibodies to high
molecular weight glutenin subunits for use in measurement of gluten in foods. F
Nutr Rep 2015; 1(2): 10-18.
9. Skerritt JH, Hill AS. Monoclonal antibody sandwich enzyme immunoassays for
determination of gluten in foods. J Agric Food Chem 1990; 38: 1771-1778.
10. Wieser H, Koehler P. Is the calculation of the gluten content by multiplying the
prolamin content by a factor of 2 valid? Eur Food Res Technol 2009; 229: 9-13.
11. Wieser H, Seilmeier W, editor. Determination of gliadin and gluten in wheat
starch by means of alcohol extraction and gel permeation chromatography.
Proceedings of the 17th Meeting of the Working Group on Prolamin Analysis and Toxicity; 2002; London, UK. Zwickau: Verlag Wissenschaftliche Scripten 2003.
12. van Eckert R, Bond J, Rawson P, et al. Reactivity of gluten detecting monoclonal
antibodies to a gliadin reference material. J Cereal Sci 2010; 51: 198-204.
13. Allred LK, Ritter BW. Recognition of gliadin and glutenin fractions in four
commercial gluten assays. J AOAC Int 2010; 93: 190-6.
14. Ciclitira PJ, Ellis HJ, Lundin KEA. Gluten-free diet - what is toxic? Best Pract Res
Clin Gastroenterol 2005; 19: 359-71.
15. Sollid LM, Qiao SW, Anderson RP, et al. Nomenclature and listing of celiac
disease relevant gluten T-cell epitopes restricted by HLA-DQ molecules.
Immunogenetics 2012; 64: 455-60.
16. Kasarda DD, Dupont FM, Vensel WH, et al. surface-associated proteins of wheat
starch granules: suitability of wheat starch for celiac patients. J Agric Food Chem
2008; 56: 10292-302.
17. Wieser H, Antes S, Seilmeier W. Quantitative determination of gluten protein
types in wheat flour by reversed-phase high-performance liquid chromatography.
Cereal Chem 1998; 75: 644-50.
Comparison of immunomethods for the characterisation
of gluten immunogenic peptides in a commercial
beer
Real2, Carolina Sousa2, María Isabel Torres3, Elena Quesada-Hernández1, Ángel
Cebolla1
1 Biomedal SL, Sevilla, Spain
2 Dpto. de Microbiología y Parasitología, Facultad de Farmacia, Universidad de
Sevilla, Sevilla, Spain
3 Dpto. de Biología Experimental, Campus Universitario Las Lagunillas, Jaén, Spain
Introduction
Gluten is present in the most commonly consumed cereals (wheat, barley, rye and
oats) and serves as ingredient in many processed foods. Manufacturing of processed
foodstuffs digests gluten to different degrees, especially by hydrolysis and
fermentation. This digestion of total gluten gives rise to peptides and, ultimately, to
amino acids. In the small intestine, some of these peptides are resistant to
gastrointestinal digestion and trigger the immune response that causes the symptoms
of the disease. This pool of peptides is termed gluten immunogenic peptides (GIP).
However, the characterization of these peptides is still incomplete. The great
heterogeneity of gluten proteins makes this task complicated and tedious. In the last
years, it has been shown that a few highly immunogenic peptides could account for
more than 90% of the coeliac-specific response [1-3]. The dominant immunogenic
peptide in wheat is the α-gliadin 33-mer [2].
Beer is the most widely consumed alcoholic beverage, both among coeliac and noncoeliac
individuals. Its production involves the fermentation of starches, mostly from
cereal grains (barley, wheat, maize, rice…). This fermentation hydrolyses gluten
proteins contained in the cereal grains and produces GIP which remain in the final
product. The differential hydrolysis of prolamins in brewing processes may generate
peptide pools with uncertain immunogenicity. Current methods based on the R5
antibody to officially analyse gluten content in beer and grant the “gluten-free” label
may overlook these immunogenic peptides. However, the new generation of
monoclonal antibodies (mAbs) like A1 and G12 with a sensitivity and specificity for
the 33-mer several orders of magnitude higher compared to R5 antibody may result in
differences in immunogenicity estimation for hydrolytic prolamins [7]. Beers, due to
their diversity, are some of the samples in which the immunomethods may show the
highest differences in gluten content measurement. Here, we evaluated the reliability
of the methods based on R5 and G12 to estimate the potential toxicity by GIP
contained in a commercial beer, which was previously characterised by HPLC-MS and
peripheral blood mononuclear cell (PBMC) reactivity from coeliac patients.
Materials and methods
Beer samples, negative control (rice prolamins), peptide synthesis, and synthetic
peptides were used. Patients with active coeliac disease and healthy subjects were
included in this study. T-cell isolation from coeliac patients, cell proliferation assays,
interferon (IFN)-γ, lateral flow immunoassay (LFIA) A1/G12 (GlutenTox® Sticks,
Biomedal), competitive ELISA G12 (GlutenTox® Competitive G12 Biomedal) and
R5 (Ridascreen, R-Biopharm), and immunoprecipitation assays were made as
described in [4,5] and are not reproduced here for space reasons.
Results and discussion
In a previous study, we characterised about 100 Belgian beers by LFIA and ELISA
based on G12/A1 mAbs (some examples showed in Table 1 and [6]). Although the
sandwich ELISA configuration may underestimate the presence of some gluten
peptides with only one epitope, LFIA and G12 competitive ELISA provided a similar
estimation of gluten content. However, the underestimation appeared to be higher in a
R5 sandwich than in the A1/G12 LFIA (Table 1). This observation may indicate that
the abundance of tandem epitopes for A1 and G12 is more frequent than that of R5
epitopes.
To analyse the differential epitope recognition present in beers, we selected a beer
based on the difference in gluten estimation by R5 and G12 ELISA (Table 1, in bold).
The Strong Ale 5 was fractionated and characterised with HPLC-MS and the gluten
content of each fraction was further analysed using LFIA A1/G12. All immunoreactive
fractions contained peptides recognised by A1, G12 and R5. Five peptides were
selected according to the presence of epitopes with potential immunogenicity (i.e.
reactive to R5, A1 and G12). These peptides were synthesised ([4] and Fig. 1). The R5
competitive ELISA showed 5- to 9-fold less reactivity for the barley beer epitopes
compared to the G12 competitive ELISA. A1 competitive ELISA showed an
intermediate affinity for the immunogenic peptides compared to R5. The biggest
differences in reactivity were found in peptide QP 22.2, which contains two tandem
epitopes for R5 and one for A1 (QP 22.2 in Fig.1). QP22.2 reactivity for R5 was sixfold
larger than to A1 and a hundred-fold larger than for G12. Interestingly, despite its
great reactivity to R5, this peptide induced a very weak reactivity to PBMCs from
coeliac patients, slightly superior to the negative control (rice prolamins) (Fig. 2 and
[4]). In contrast, the most reactive peptide for G12 (PP 24.1) also confirmed the
highest immunogenicity by PBMC activation and IFN-γ production. These results
were consistent with those obtained by G12/A1 competitive ELISA, but not according
to R5. Therefore, there is no correlation between the reactivity for the R5 mAb and the
immunogenicity of peptides. Moreover, the highest sensitivity of G12 for such GIP
could be an indication of the presence of immunogenicity risks in many cases.
Table 1. Gluten levels of the 30 Belgian beer samples analysed with mAbs G12/A1 and
mAb R5 ELISA.

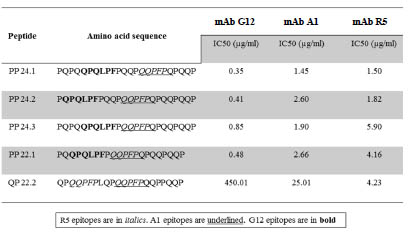
Figure 1. Relative affinity of G12, A1 and R5 mAbs for different immunoreactive
peptides from the barley beer previously characterised by HPLC-MS and PBMC
activation.
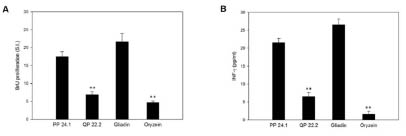
Figure 2. Potential immunogenicity of PP 24.1 and QP 22.2. (A). Proliferative
responses of PBMCs to different peptides. (B). IFN-γ production by PBMCs with
different peptides. Results are expressed as mean ± SD of duplicated cultures (n = 14).
Gliadin and oryzein were used as the positive and negative control, respectively, and
significant differences with respect to gliadin at **p<0.005 are shown
Next, we wanted to assess the similarities between the pool of GIP detectable by G12
and those detectable by R5. To do so, we fractionated the Strong Ale 5 beer with
immunochromatography using agarose beads conjugated with G12 mAb (Fig. 3).
Three fractions were obtained: Input (barley beer), Flow Through (barley beer minus
the G12 reactive peptides) and Output (peptides bound to the G12 columns that are
released by heat denaturation). All three fractions were characterised and we quantified the relative content of proteins, G12 and R5 reactive species, and T-cell
activation.

Figure 3. Relative content of proteins, G12 and R5 reactive peptides and coeliac
immunogenic pattern of the different barley beer fractions. Fractions were obtained in
the process of separation of peptides by G12 mAb immunodepletion
As expected, material immunocaptured by G12 (peptides and proteins) comprised no
more than 5% of the total protein content in the beer. However, these peptides were
responsible for about 90% of the immunogenicity of the total beer. Strikingly, almost
80% of the reactivity of the R5 mAb is located in the pool of peptides and proteins
with poor immunogenic activity, as corroborated by the proliferative responses of Tcells
and IFN-γ production by PBMCs of coeliac patients.
Conclusions
We have shown that analysing a beer with standard methods like ELISA R5 to grant it
the “gluten-free” label might not fully guarantee the absence of potential damage to
coeliac patients. There is experimental evidence that a better indicator of the potential
immunogenicity is the reactivity to the -gliadin 33-mer A1 or G12, even if the 33-
mer canonical sequence is not supposed to be present in barley. Other epitopes of the
33-mer are enough to sensitively detect GIP by G12/A1 in barley beers.
In contrast, the presence of the preferred R5 epitope QQPFP in certain peptides of the
Strong Ale 5, was not sufficient to detect those GIP that appeared detectable by
HPLC-MS analysis. In general, the G12 immunomethods appear to be the most specific practical techniques described so far to assess the potential immunogenicity of
barley beers.
References
1. Anderson RP, Degano P, Godkin AJ, et al. In vivo antigen challenge in celiac
disease identifies a single transglutaminase-modified peptide as the dominant Agliadin
T-cell epitope. Nat Med 2000; 6: 337-42.
2. Shan L, Molberg Ø, Parrot I, et al. Structural basis for gluten intolerance in celiac
sprue. Science. 2002; 297: 2275-9.
3. Tye-Din JA, Stewart JA, Dromey JA, et al. Comprehensive, quantitative mapping
of T cell epitopes in gluten in celiac disease. Sci Transl Med 2010; 2: 41-51.
4. Real A, Comino I, Moreno Mde L, et al. Identification and in vitro reactivity of
celiac immunoactive peptides in an apparent gluten-free beer. PLoS One 2014;
9:e100917.
5. Moreno Mde L, Muñoz-Suano A, López-Casado MÁ, et al. Selective capture of
most celiac immunogenic peptides from hydrolyzed gluten proteins. Food Chem
2016; 205:36-42.
6. Comino I, Real A, Moreno Mde L, et al. Immunological determination of gliadin
33-mer equivalent peptides in beers as a specific and practical analytical method to
assess safety for celiac patients. J Sci Food Agric 2013; 4: 933-43.
7. Torgler C, Síglez MA, Vílchez F et al. Analytical tools to detect gluten
immunotoxic fractions in food based on monoclonal antibodies raised against the
gliadin 33-mer peptide. 24th Proceedings of the WGPAT 2011.
Pathogenesis of coeliac disease: complexes between
transglutaminase and gluten peptides
Barbara Lexhaller, Peter Koehler, Katharina A. Scherf
Deutsche Forschungsanstalt für Lebensmittelchemie, Leibniz Institut, Freising,
Germany
Introduction
Coeliac disease can be characterised by three features: (A) triggered by the ingestion
of gluten, (B) presence of the genetic factor (HLA-DQ2 or DQ8), and (C) the
generation of autoantibodies against tissue transglutaminase (TG2) [1]. After the
ingestion of gluten, these proteins (gliadins, glutenins, hordeins, and secalins) are not
sufficiently digested by human gastrointestinal enzymes due to their high proline and
glutamine contents. These long peptides pass through the epithelial layer and first
trigger the innate immune response. Intraepithelial lymphocytes activate defence
mechanisms, which initiate apoptosis and are increase of epithelial permeability.
Secondly, the gluten peptides are modified by TG2 that catalyses deamidation and
transamidation. The modified peptides stimulate gluten-specific T-lymphocytes, which
finally lead to the damage of the villi of the small intestine. Furthermore, antibodies
are formed against gluten peptides, TG2 and gluten peptide-TG2-complexes [1-3].
TG2 plays a key role in the pathogenesis of coeliac disease. Firstly, it causes
deamidation of specific glutamine residues to glutamic acid, which increases the
immune response. It also initiates transamidation and formation of gluten peptide-
TG2-complexes that lead to the formation of antibodies against them. TG2 is a Ca2+-
dependent protein-glutamine γ-glutamyltransferase (EC 2.3.2.13), which catalyses the
formation of inter- and intramolecular Nε(γ-glutamyl)lysine bonds. The transfer of the
acyl residue between the -carboxyamine group of glutamine as acyl donor and
primary amines as acyl acceptors involves a two-step reaction mechanism. The three
amino acids cysteine-277, histidine-335, and aspartic acid-358 of the active site of the
enzyme are involved in this mechanism. According to the hypothetical model of “hapten-carrier-like complexes” these covalently bound gluten peptide-TG2-
complexes should be responsible for the formation of anti-TG2 antibodies.
However, the investigation of the structures of these gluten peptide-TG2-complexes is
still at the beginning. Therefore, the aim of this study was to identify the binding sites
between TG2 and peptides derived from all CD-active gluten protein types of wheat,
rye, and barley.
Materials and methods
Characterisation of microbial transglutaminase
The microbial transglutaminase from Streptomyces mobaraensis (ABEnzymes,
Darmstadt, Germany) was dissolved in formic acid (0.1%), filtered (0.45 μm) and
measured by liquid chromatography with mass spectrometric detection (LC-MS
(QTOF)). Furthermore, the microbial transglutaminase was characterised by the
analysis of the tryptic peptides by LC-MS/MS (iontrap). For this purpose, the enzyme
was incubated with trypsin in TRIS-HCl-buffer (0.1 mol/l; pH 7.8) for 24 h at 37 °C.
After purification with solid phase extraction, the hydrolysates were dried, dissolved
again in formic acid (0.1%) and analysed by LC-MS/MS (iontrap).
Identification of isopeptides
For the reaction of microbial transglutaminase and a defined model peptide gli 56-75
(LQLQPFPQPQ65LPYPQPQLPY) to peptide-enzyme-complexes, both were dissolved
in TRIS-HCl-buffer (0.1 mol/l; pH 7.8; 2 mmol/l CaCl2) and incubated for 2 h at
37 °C. The peptide-enzyme-complexes were incubated with trypsin for 24 h at 37 °C.
After purification with solid phase extraction the hydrolysates were analysed by LCMS/
MS (iontrap).
Results and discussion
Characterisation of microbial transglutaminase
Initially, the microbial transglutaminase (mTG) had to be characterised by molecular
weight and by sequence analysis. The characterisation by molecular weight was
carried out by LC-MS/MS (QTOF) with a high intensity. The identified molecular
weight of the microbial transglutaminase was determined as 37,863.6 ± 0.5 (Fig. 1),
which is comparable to the data (P81453) of the UniProt KB database. Also Kanaji et
al. could identify the same molecular weight for microbial transglutaminase and
showed the separation of signal- and propeptide during the MS measurement [4].

Figure 1. Mass spectrum of microbial transglutaminase. The spectrum corresponds to
the average of scans of the base peak-chromatogram at 6.7-7.1 min. The simulated
maximum entropy peak is shown in the upper right corner
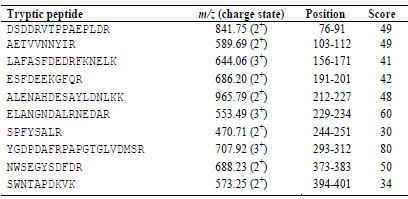
Furthermore, the analysis of the tryptic peptides of mTG confirmed the
characterisation and comparability with P81453. Thirteen tryptic peptides were evenly
distributed over the whole sequence, without the signal- and propeptide.
Table 1. Tryptic peptides of microbial transglutaminase, their m/z ratio with charge
state, the position in the UniProt database sequence and the score of the search with
MASCOT-software (≥ 30).
Identification of isopeptides
To identify the binding sites between mTG and the model peptide, the
transglutaminase was first allowed to react with the peptide, the complexes were
hydrolysed and these tryptic peptides were analysed with LC-MS/MS. In the second
part, an analysis strategy for the identification of the isopeptides had to be developed.
For this purpose, all theoretically possible combinations between a lysine residue (K)
of the enzyme and a glutamine residue (Q) of the model peptide, their masses and their
precursors (m/z) in the different charge states were calculated and theoretically
fragmented. In the next step, the measured MS- and MS/MS-spectra were searched for
the calculated masses of the precursors and fragments.
For the identification of the binding Q, a data analysis strategy had to be developed to
detect fragments which confirm the binding site. Initially, the spectra were searched
for isopeptide bonds at Q65, because Fleckenstein et al. (2004) already showed that this
position is a binding site for TG2 [5]. Fig. 2 presents the isopeptide of the model
peptide with the possible binding site (Q65) and the tryptic peptide KWQQVYSHR of the
transglutaminase. To confirm the position of the isopeptide bond at Q65 the specified
fragments b10α or b11α as well as b9α, b8α, y11α or y12α and y10α had to be identified. At
last, to confirm the identification of the whole isopeptide, 5 other fragments, e.g. b5β or
y6β had to be identified.
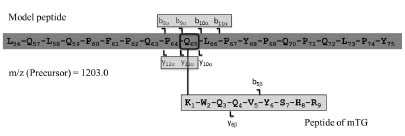
Figure 2. Strategy for identification of the binding Q in isopeptide bonds. Isopeptide
(precursor m/z = 1203.0 (3+)) of the model peptide and the tryptic peptide
KWQQVYSHR of microbial transglutaminase (mTG) with the specified fragments for
confirmation of the binding site
The mass spectrum of the isopeptide of the model peptide gli 56-75 and the tryptic
peptide KWQQVYSHR of the microbial transglutaminase is presented in Fig. 3.
Twenty-three fragments of the b- and y-series could be identified in the entire
isopeptide and all signals had an adequate intensity. Another condition for the certain
identification is to identify at least three consecutive fragments. With the detected
fragments b10α to b14α and b5β to b8β as well as y8α to y10α and y12α to y14α this
requirement was fulfilled. Also the conditions for the affirmation of the binding Q were fulfilled by the identification of the specified fragments b10α and b11α as well as
y12α and y10α.
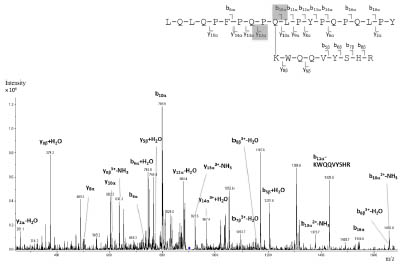
Figure 3. Mass spectrum of the isopeptide (m/z = 1203.0 (3+)) of the model peptide gli
56-75 and a tryptic peptide of the transglutaminase
Until now, five tryptic peptides of mTG were identified, which form isopeptide bonds
with the Q65 of the model peptide gli 56-75. Tab. 2 shows these tryptic peptides and
their position in the transglutaminase sequence.
Table 2. Identified tryptic peptides of the microbial transglutaminase, which bind to
the model peptide gli 56-75 at Q65, their position in the sequence and m/z ratio and
the charges state of the formed isopeptides.

Conclusions
First, the characterisation of the microbial transglutaminase by molecular weight and
sequence analysis was performed by two different types of LC-MS/MS. The results of
the microbial transglutaminase used here were comparable with the data of the UniProt
KB database (P81453).
The preliminary experiments on the identification of isopeptides were focused on the
development of a strategy, whereby first results were achieved. Until now, this
strategy allowed the identification of five isopeptides with the binding site at Q65 of the
model peptide gli 56-75.
References
1. Schuppan D, Junker Y, Barisani D. Celiac disease: from pathogenesis to novel
therapies. Gastroenterology 2009; 137: 1912-1933.
2. Kagnoff M. Celiac disease: pathogenesis of a model immunogenetic disease. J
Clin Invest 2007; 117: 41-49.
3. Qiao SW, Iversen R, Ráki M et al. The adaptive immune response in celiac
disease. Semin Immunopathol 2012; 34: 523-540.
4. Kanaji T, Ozaki H, Takao T et al. Primary structure of microbial transglutaminase
from Streptoverticillium sp. strain. J Biol Chem 1993; 268: 11565-11572.
5. Fleckenstein B, Qiao SW, Larsen MR et al. Molecular characterization of
covalent complexes between tissue transglutaminase and gliadin peptides. J Biol
Chem 2004; 279: 17607-17616.
Potential of non-prolamin storage proteins in coeliac
disease
Gyöngyvér Gell1, Gábor Veres2, Ilma Rita Korponay-Szabó3, Angéla Juhász1
1 Department of Applied Genomics, Agricultural Institute, MTA Centre for Agricultural
Research, Martonvásár, Hungary
2 First Department of Paediatrics, Semmelweis University of Medicine, Budapest,
Hungary
3 Celiac Disease Centre, Pál Heim Children's Hospital, Department of Paediatrics,
Budapest, Hungary
Introduction
Brachypodium distachyon is a small annual grass that belongs to the Pooideae
subfamily of the grasses, and based on the recent phylogenetic analyses is the closest
wild relative of wheat and barley. This wild grass with special biological properties
(small size, rapid generation time and self-fertility) and genomic attributes (small
genome (272Mbp), diploid accessions) is suitable for use as a model system of cereals.
B. distachyon accession Bd21 offers many advantages, such as self-fertility, simple
nutrient requirements and short lifecycle. Sequencing and annotation of the Bd21
genome were recently completed, making further functional proteomic studies feasible
[1-3]. The main storage proteins of Bd21 are the 11S, 12S and 7S globulin type
proteins similar to oat and rice. The prolamins, including the avenin-like proteins and
the gliadin-like prolamins, represent less than 12% of the total protein content which is
significantly lower compared to wheat or barley. Due to specific proteomic features
this annual grass is a good model plant to investigate the toxic nature of non-prolamin
seed storage proteins. 11S-12S globulins account for 70-80% of total seed protein
content [4]. In our previous study, the published chromosome specific B. distachyon
genome sequences and a seed specific cDNA library data were used for sequencebased
identification of proteins with regions identical to known coeliac diseasespecific
epitopes [5]. These results have highlighted the presence of possible crossreactive
epitope homologues to coeliac disease-related trigger molecules. Although
Brachypodium is not considered for human nutrition, we took advantage of its use as a
model species for the understanding whether abundant non-prolamin cereal seed
proteins with linear epitope homologues play a role in the development of the humoral
immune response and to help select other food sources suitable for a gluten-free diet.
Materials and methods
Serum samples from coeliac patients with known HLA-DQ haplotypes and positive
for coeliac disease antibodies on gluten intake (n = 13, 8 females, 5 males), median
age 5.7 years, range 1.4 - 13.5 years), serum samples from coeliac patients adhering to a strict gluten-free diet (GFD) resulting in normalised antibodies (antitransglutaminase
IgA < 10 U/l) and mucosal healing (n=3), from ten newly diagnosed
Crohn’s disease patients with ileocolon manifestation (median age 6.4) and from eight
healthy control subjects were used for immunoblotting studies.
In case of Bd21 total protein extracts, proteins were extracted with SDS buffer
following the protocol of Dupont and co-workers [6] which extracted a greater
percentage of protein from wheat flour than other methods and facilitated removal of
starch.
After the 2D gel electrophoresis (GE) the proteins were transferred to an ImmobilonP
PVDF membrane and IgA-based immunoblots were carried out. Rice glutelin antibody
coupled with anti-rabbit IgA as a secondary antibody was used in 2D Western blot
analysis to confirm the presence of seed storage globulins [7].
Protein sequences identified based on the online nanoLC-MS/MS analysis were
retrieved from the UniProt database and were used for detailed epitope mapping
analyses and protein characterisations. p-BLAST was used to find protein homologues
in Poaceae. Coeliac disease-specific linear T-cell and B-cell epitopes were collected
from the ProPepper database [8]. Epitope mapping was carried out using the motif
search algorithm of the CLC Genomic Workbench (8.5.1).
Results and discussion
Brachypodium distachyon, a model plant of monocot species with low prolamin
content was investigated to characterise immune reactivity against non-prolamin
proteins in the seed. Altogether, 28 immune-reactive protein spots were analysed by
online nanoLC-MS/MS (Fig 1.). Antibody reactivity against Brachypodium proteins
was detected in all coeliac disease patients and two of Crohn’s disease patients. While
positive IgA reactions of coeliac serum samples were detected against proteins from a
wide range of molecular weight (approximately 15,000 to 65,000) and variable
isoelectric points, the protein spots showing immune reactivity with Crohn’s disease
serum samples possessed an approximate molecular weight of 24,000. No proteins
reacted when sera of healthy controls and sera of patients on a strict gluten-free diet
were applied. Most of the spots were identified as 7S or 11S-12S type seed storage
globulins. A few prolamin-type storage protein sequence hits were also identified as
secondary or tertiary protein hits with similar sequence coverage values: gamma
gliadin-like proteins, HMW glutenin-like proteins, and LMW type respectively. Some
enzymes and proteins with non-storage function were identified in the 35,000 region,
like glucose/ribitol dehydrogenase, aldo-keto reductase, xyloglucan endotransglucosylase/
hydrolase and aspartic peptidase.
The most significant globulin hits (I1GPS5, I1GMC8, I1HMK7, I1IPF2 I1HNH9) as
well as the most frequently identified prolamin protein (I1HRM6) were subjected to in
silico sequence analyses. Two adjacent cupin-1 domains characteristic for both 7S and 11S-12S seed storage globulins were found in all of the Brachypodium globulin hits (Fig. 2). The identified epitope homologues represented peptides with polyQ stretches
and were positioned in two glutamine-rich regions of the protein. Additionally, a
peptide with six residues (QPEQPF) was identified in the 11S seed storage globulin
I1HNH9. This peptide was the deamidated version of a known immune reactive AGAspecific
B-cell epitope QPQQPF (IEDB Epitope ID 147232) that gets deamidated
during coeliac disease pathogenesis. This deamidated version represents one of the
primary targets of serum deamidated gliadin peptide (DGP) antibodies in coeliac
disease. Interestingly, none of the I1HNH9 Poaceae homologues contained this
deamidated peptide. No epitope homologues were found in the metabolism-related
proteins.
Figure 1. Identification of coeliac disease-related proteins of Brachypodium
distachyon ‘Bd21’ using anti-IgA detection and patients’ blood sera. (A) 2D gel
electrophoresis of total protein extract of inbred line Bd21. Proteins were separated
on 3-10 pH IPG strips followed by separation on 12% acrylamide gels. Labelled
protein spots represent immune-reactive proteins and were sent for online nanoLCMS/
MS analyses. Molecular weight range is marked on the left-hand side. (B) and (C)
Representative immunoblots using sera with IgA reactivity of therapy naïve coeliac
disease patients, (B) HLA-DQ2, (C) HLA-DQ8
When gluten-related known T-cell epitopes were mapped to the Brachypodium
proteins, no known epitopes were found. However, a type-I diabetes-specific T-cell
epitope, EEQLRELRRQ [9] was identified from I1GPS5 with 100% sequence identity
at the position 281. To check for novel T-cell epitopes, MHC-II binding predictions
were carried out from the main 7S, and 11S-12S globulin hits using HLA-DR3-DQ2
and HLA-DR4-DQ8 MHC-II haplotypes (Fig. 2.) [10]. Some previous studies have
focused on the investigation of immune reactivity and toxic behaviour of non-gluten
proteins of wheat related to coeliac disease [11-14]. Recently, Huebener and
colleagues have analysed the possible involvement of non-gluten proteins as target
antigens in coeliac disease-related humoral response [15]. Serine-protease inhibitors,
alpha-amylase inhibitors, farinins and seed globulins have demonstrated a significant
immune response. Additionally, 35% of coeliac disease patients’ sera showed
reactivity against protein spots identified as seed globulins using the protein extract of ‘Butte86’ wheat [14]. Increased coeliac serum antibody reaction was also measured
against cereal globulin extracts by Troncone et al. [13]. Although 7S and 11S-12S seed
storage globulins both represent strongly conserved protein families with cupin-1
domains in cereals, our epitope analyses highlighted some remarkable differences
between the protein families [10]. These differences indicate the presence of possible
sub-classes with various immune-reactive potential. The amount of these unique
globulin sub-classes can also be different in the grains of the different species, with a significantly lower amount expressed in wheat and in cereals, where prolamins serve
as major storage protein components. This fact partially explains why these proteins
were overlooked compared to the most abundant prolamins in wheat, rye or barley.
Our study confirmed that globulin-type cereal seed storage proteins are specifically
related to coeliac disease, as patients suffering from other immunological
inflammatory diseases, like Crohn’s disease did not recognise these globulin-type
cereal storage proteins. Adverse results of the immunoblot analyses with sera of
coeliac patients on a gluten-free diet had also strengthened the assumption that seed
storage globulins may act as secondary B-cell stimulants due their strong sequence
homology to epitopes originated from the primary gluten triggers. In progressive
stages of the disease, villous atrophy and increased gut permeability contribute to why
these proteins can serve as cross-antigens. The recovered intestinal mucosa of the
coeliac patient on a strict gluten-free diet better prevents the passage of ingested
proteins and probably, in this way, the strong immune reactivity can be controlled.
Figure 2. MHC II class T-cell epitope prediction of Brachypodium 7S and 11S-12S
globulin proteins using HLA-DQ2 and -DQ8 alleles and IEDB analysis resource
Consensus tool. Selection of predicted binders was carried out using the top 1%
binders based on consensus percentile rank values. Predictions were calculated for
each allele separately. Predicted epitopes are mapped to the protein sequences
Conclusion
In summary, our results indicate that both in coeliac disease and type-I diabetes MHCII-
presentation and B-cell response may be developed not only for prolamins, but also
for seed storage globulins even in distant relatives of wheat, such as Brachypodium
distachyon, having seed storage globulins similar to oat and rice. However, its seed
storage proteins, especially globulins, belong to quite conserved proteins in plants,
which when eaten, may cause some problems due to the presence of some B-cell
epitope homologues and possible T-cell reactive peptides present in the globulin
fraction. Therefore, such cereals would not be harmless food alternatives for coeliac
patients. High-resolution 2D gel electrophoresis followed by immunoblotting and
protein identification have proven that 7S and 11S-12S seed storage globulins may act
as antigens for coeliac disease specific IgA antibodies. Storage globulins are only
present as contaminants in wheat gluten; therefore they play a less significant role.
Contrary to this, seed storage globulins are the main source of nutrient storage in
cereals like rice, oat or Brachypodium. Despite the strongly conserved structure of 7S
globulins, proteins like Glo3A in wheat and I1GPS5 in Brachypodium and some of the
11S-12S globulins represent a special class of seed globulins with an epitope-dense
region between the two cupin-1 domains and therefore might represent a higher risk
for coeliac disease patients. Our study draws attention on the presence of conserved
seed storage protein families in various cereal species, such as wheat, oat, rice and
Brachypodium. Although 7S and 11S-12S seed globulins are present in low amounts
in the wheat grain, they represent major storage protein groups in species like oat or
rice. Therefore the presence of some epitopes in highly conserved regions may be also
characteristic on orthologues in other species. The level of the response to globulins
may depend on the type of seed globulins and amount of proteins with immune
responsive peptide content. It is also suggested to perform further investigations
whether diets enriched in seed storage globulins (like rice or oat) inhibit sufficient healing, especially in patients with combined high risk to type-I diabetes and proven
susceptibility to these proteins.
Acknowledgement
This project was supported by the European Union and co-financed by the European
Social Fund (grant agreement no. TÁMOP-4.2.2.A-11/1/KONV-2012-0008), OTKA
101788, OTKA K10088 and OTKA PD 115641.
References
1. Ozdemir BS, Hernandez P, Filiz E, et al. Brachypodium genomics. Int J Plant
Genomics 2008; 2008: 536104.
2. Peraldi A, Beccari G, Steed A, et al. Brachypodium distachyon: a new
pathosystem to study Fusarium head blight and other Fusarium diseases of wheat.
BMC Plant Biol 2011; 11: 100.
3. Hands P, Drea S. A comparative view of grain development in Brachypodium
distachyon. J Cereal Sci, 2012; 56: 2-8.
4. Larré C, Penninck S, Bouchet B, et al. Brachypodium distachyon grain:
identification and subcellular localization of storage proteins. J Exp Bot 2010;
61(6): 1771-1783.
5. Juhász A, Gell G, Sebestyén E, et al. Brachypodium distachyon as a model for
defining the allergen potential of non-prolamin proteins. Funct Integr Genomics
2012; 12: 439-446.
6. Dupont FM, Vensel WH, Tanaka CK, et al. Deciphering the complexities of the
wheat flour proteome using quantitative two-dimensional electrophoresis, three
proteases and tandem mass spectrometry. Proteome Sci 2011; 9 (10): 1-29.
7. Krishnan HB, Okita TW. Structural relationship among the rice glutelin
polypeptides. Plant Physiol 1986; 81: 748-753.
8. Juhász A, Haraszi R, Maulis C, et al. ProPepper: a curated database for
identification and analysis of peptide and immune-responsive epitope composition
of cereal grain protein families. Database 2015; article ID bav100,
doi:10.1093/database/bav100.
9. Scott LJ, Mohlke KL, Bonnycastle LL, et al. A genome-wide association study of
type 2 diabetes in Finns detects multiple susceptibility variants. Science 2007; 316
(5829): 1341-5.
10. Gell G, Kovács K, Veres G, et al. Characterization of globulin storage proteins of
a low prolamin cereal species in relation to celiac disease. Sci Rep 2017; 7: 39876.
11. Stern M, Fischer K, Gruttner R. Immunofluorescent serum gliadin antibodies in
children with coeliac disease and various malabsorptive disorders. II. Specificity of gliadin antibodies: immunoglobulin classes, immunogenic properties of wheat
protein fractions, and pathogenic significance of food antibodies in celiac disease.
Eur J Pediatr 1979; 130(3): 165-172.
12. Kieffer M, Frazier PJ, Daniels NW, et al. Wheat gliadin fractions and other cereal
antigens reactive with antibodies in the sera of celiac patients. Clin Exp Immunol
1982; 50(3): 651-660.
13. Troncone R, Auricchio S, De Vincenzi M, et al. An analysis of cereals that react
with serum antibodies in patients with coeliac disease. J Pediatr Gastroenterol
Nutr 1987; 6(3): 346-350.
14. Penttila IA, Devery JM, Gibson CE, et al. Cellular and humoral responses in
coeliac disease. 1. Wheat protein fractions. Clin Chim Acta 1991; 204(1-3): 95-
107.
15. Huebener S, Tanaka CK, Uhde M, et al. Specific nongluten proteins of wheat are
novel target antigens in celiac disease humoral response. J Proteome Res 2015;
14(1): 503-511.
Preview of the ‘Well on Wheat?’ (WoW) project
Twan AHP America1, Luud JWJ Gilissen1, Marinus JM Smulders1, Peter Shewry2,
Flip van Straaten3, Daisy Jonkers4, Fred Brouns4
1 Wageningen University & Research, Wageningen, The Netherlands
2 Rothamsted Research, Harpenden, United Kingdom
3 Dutch Bakery Institute, Wageningen, The Netherlands
4 Maastricht University, Maastricht, The Netherlands
Abstract
The ‘Well on Wheat’ (WoW) project aims to generate robust data on the effects of
wheat-based food products on gastrointestinal function and metabolism. The first
objective of the project is to obtain in-depth analytical data of the composition of
whole meal obtained from bread wheat, spelt wheat and emmer wheat as well as the
dough and the finally baked bread made thereof. The second objective is to study the
effects of two alternative fermentation processes: yeast and sourdough fermentation,
on compositional changes. Objective 1 and 2 will give insight in the overall effects of
food processing on the (bio)chemical composition as defined by proteomics,
carbohydrate analysis (carbohydrates, fibers, FODMAPs), phytate, selected
micronutrients and pesticide residues. The third objective is to study the effects of
consuming the various bread types (according to grain type and fermentation type) in
individuals with irritable bowel syndrome (IBS), which will be monitored for effects
on intestinal function and physiology, including e.g. faecal microbiota/metabolism,
and using markers for gut permeability and inflammation as well as measuring
subjective perceptions. The project will generate new scientific insights that will be
translated into recommendations to food industries, health professionals and
patient/consumer organisations. This project will be carried out in the framework of
the Health Grain Forum and supported by the ICC (International Association for
Cereal Science and Technology) and funded by private and public organisations.
Introduction
During the last decade, a significant movement to the adoption of gluten-free and
wheat-free foods has developed in Western societies. The prevalence of wheat
intolerance (coeliac disease) and wheat allergy are well known, being 1% and 0.2%
of the general population, respectively. However, in the US, nearly 30% of the adult
population has expressed a desire to reduce or eliminate wheat and/or gluten from their
daily diet [1] while a recent questionnaire-based study in the Netherlands [2] showed
that 6.2% of a cohort of 785 adults reported adverse symptoms after the ingestion of
gluten-containing foods. The most widely reported intestinal symptoms were bloating,
abdominal discomfort and flatulence, but extraintestinal symptoms were also mentioned such as fatigue and headache. Symptoms were generally experienced
several days a week, starting mostly between one and six hours after consumption and
lasting several hours. These self-reporting ‘gluten sensitive’ individuals were mainly
younger females (80%) living in urban regions with a trend of higher education
levels (which confirms previous data of a UK study on self-reported gluten sensitivity
[3]). Over one third of the reported symptoms met the consensus criteria for a positive
diagnosis, the ‘Rome III criteria for IBS’, which have been established due to the
absence of reliable biomarkers and specific laboratory tests [4].
The reasons why so many people feel more comfortable on a gluten-free diet may
extend beyond the food itself. Several popular books [5-7] and many statements on
social media have promoted gluten-free (‘Palaeolithic’) diets, suggesting that wheat
consumption has adverse health effects leading to various chronic diseases.
Furthermore, it is often claimed that products made from modern bread wheat varieties
have negative health effects, but not foods made from so-called ‘ancient’ wheats such
as spelt (which is closely related to modern bread wheat) and emmer (which is more
closely related to modern pasta wheat), which are generally cultivated under organic
conditions. These messages are, however, in contradiction to ample scientific data that
have demonstrated significant health-promoting effects of whole grain consumption
[8-12]. Despite these proven health benefits, the negative messages have resulted in a
significant decline in the consumption of breads and other wheat products in Western
countries.
In this context, IBS has often been considered by the patients themselves to be
associated with food and especially wheat consumption. IBS is the most commonly
diagnosed functional gastrointestinal (GI) disorder with a prevalence of 10-20%
worldwide, predominantly among women [13]. Structural abnormalities and tissue
damage are generally absent, but psychiatric co-morbidity is often reported, indicating
a psychosomatic component in a subgroup of these patients. Although several factors
have been associated with IBS, including e.g. microbial perturbations, altered
permeability, motility and visceroperception, the exact pathophysiology is not yet
clear. Also markers for mucosal immune activation and inflammatory responses have
been reported in a subset of IBS patients that may disappear after elimination of
wheat/gluten from the diet [14,15]. This condition is often referred to as ‘non-celiac
wheat sensitivity’ (NCWS) or ‘non-celiac gluten sensitivity’ (NCGS) [16].
Dietary factors such as FODMAPs (fermentable, oligo-, di-, monosaccharides and
polyols) have been recognised as triggers for symptoms in some subjects, by providing
substrates for colonic fermentation [16 and refs therein, 17]. It has also been reported
that replacing a bread wheat-based diet by whole grain products from ‘ancient’ wheats
such as spelt, has benefits for IBS patients [18,19]. In addition to gluten and
FODMAPs, the presence of relatively high quantities of amylase-trypsin inhibitors
(ATIs) in bread wheat has also been suggested as a potential IBS causing factor
[20,21]. Direct comparative data about the effects of foods obtained from different wheat types and their possible contribution to the pathophysiology of NCWS are,
however, still lacking. Here we propose a research strategy to address this issue.
Project design
The WoW project will study the effect of different grains in IBS patients to provide
information on the wheat- and disease-related issues at three levels: (1) The
biochemical composition of wheat grains and changes during processing steps
(milling, fermentation, baking) into consumable food products (bread); (2) The impact
of bread consumption on well-being and GI symptoms, gut permeability, immune
function and the microbiome; and (3) The impact of the opinions and perception of
consumers/patients on wheat consumption or avoidance regarding gastrointestinal
symptoms and well-being. The project will be managed by the academic and funding
partners in a contractually agreed pre-competitive manner.
Materials and methods
Grains. Grains from bread wheat (Triticum aestivum) (representing current bread
products), spelt wheat (T. aestivum ssp spelta) and emmer wheat (T. dicoccum) (both
representing ‘ancient’ wheat species) obtained commercially will be analysed for
biochemically for proteins using proteomics (detection of gluten, globulins, albumins,
ATIs, lectins, indigestible peptides), fibre (including fructans and other FODMAPs),
phytate, phenolics, minerals (such as zinc and magnesium), at the level of flours,
fermented (yeast and sourdough) doughs, and breads.
Cohort and intervention groups. We aim at measuring the effects of wheat
consumption in IBS patients, recruited from a large cohort of well-characterised IBS
patients [22] that has been established at Maastricht University Medical Centre. Three
groups will be used in the intervention study, including successively: a running-in
period (1 week), a free-from diet (2 weeks), a yeast or sourdough bread food challenge
(2 weeks), a free-from wash-out diet (2 weeks) and a sourdough or yeast bread food
challenge (2 weeks). It should be noted that the first and the second challenge are
reversed regarding the yeast and the sourdough breads. The three groups will differ in
their challenge: the groups 1, 2 and 3 will be challenged blinded with yeast and
sourdough bread from either bread wheat, spelt wheat or emmer wheat.
Sampling human materials. At each step in the challenge sequence, patient samples
will be taken from (1) the stool to analyse microbiota composition and metabolites
from bacterial protein and carbohydrate fermentation ((i.e. short chain fatty acids,
branched chain fatty acids, etc.); (2) breath metabolome to identify volatile organic
compounds reflecting host and microbial metabolism; (3) blood to determine
alkylresorcinols, inflammation markers (C-reactive protein and cytokines); zonulin;
and (4) urine sugar ratios as proxy for gut permeability. Furthermore, validated scores
will be applied to measure wellbeing and GI symptoms.
Yeast versus sourdough fermentation. Significant differences are expected in the
biochemical composition after yeast fermentation as compared to sourdough
fermentation [23]. If the results are not significant, the intervention schedule will be
adapted accordingly.
Nocebo effects. In healthy consumers, nocebo effects related to wheat/gluten
avoidance will be determined through a food challenge with a single bread type that
will be differently labelled and offered in four categories of emotional perceptivity and
acceptability.
Ethics. Before starting, the project will be evaluated, commented and approved by a
Medical-Ethical Committee.
Wheat cultivation. In an extension to the WoW project, the various wheat types (see
Grains above) will be compared after growth under different conditions (organic vs
standard) to determine effects of environment on composition. This data will may help
to explain possible differences, between the grain types obtained from different
European countries and cultivation practices.
Partners and Sponsors
The project proposal has been initiated by Maastricht University and further elaborated
together with Wageningen University & Research, the Dutch Bakery Centre (all from
The Netherlands) and Rothamsted Research (UK). Most research partner organizations
are members of Health Grain Forum (HGF) and the project fits into the activities of
the HGF working group on ‘Cereals and health’ and has been included in the general
HGF programme. The International Association for Cereal Science and Technology
(ICC; Austria) will serve as financial administrative partner for the following
sponsoring entities:
AB-Mauri Bakery Ingredients, Made, Netherlands
CSM Innovation Bakery Center, Bingen, Germany
CYMMIT, Texcoco, Mexico
DSM Food Specialties, Delft, Netherlands
Fazer Bakeries, Helsinki, Finland
ICC- Intl. Association for Cereal Science and Technology, Vienna, Austria
IWGA- Intl. Wheat Gluten Association, Kansas, USA
Lantmännen EK, Stockholm, Sweden
Mondelez, Saclay, France
Dutch Bakery Center, Wageningen, Netherlands
Nutrition et Sante, Revel, France
Puratos BV, Groot Bijgaarden, Belgium
Sonneveld Group BV, Papendrecht, Nederlands,
Zeelandia Zierikzee, Netherlands
Baking Industry Research Trust Howick, Auckland, New Zealand
Health Grain Forum, Vienna
The project is in part publicly funded by the Dutch Topsector Agri&Food.
The sponsoring organisations have neither a role in the design and execution of the
project, nor in the collection, analyses, interpretation and publication of the data.
Expected outcomes
The project will generate comparative data on the biochemical composition of grains
of bread wheat, spelt and emmer, with a major focus on those compounds expected to
have positive or negative effects on health, such as carbohydrates (starch, fibre,
FODMAPs), proteins (gluten, CD-immunogenic gluten peptides, albumins, globulins,
lectins, ATIs), phenolic compounds, phytate, and minerals (Zn; Mg). In an extension
of the project, biochemical analyses will be carried out on grains grown under different
conditions.
After milling, breads will be made using yeast fermentation and sourdough
fermentation and the compositions of the doughs and flours compared. A great
challenge is the production of breads from flours of the different grain types that are
visually and organoleptically similar, so that they can be used in the double-blind food
challenges.
Administration of the baked products to patients with IBS (i.c. NCWS) will reveal any
effects on the aetiology of non-celiac wheat sensitivity.
The project will also provide insight into the occurrence of nocebo effects by
collecting data obtained about post-consumption gastrointestinal symptoms from a
group of healthy volunteers that prefer to avoid gluten.
The results will be published in international scientific journals. New scientific
insights will be translated into recommendations to wheat-processing industries,
governmental regulatory bodies, health professionals, patients and consumers, and will
underpin innovation in the production of wheat-based foods.
Acknowledgements
Here, we would like to mention the names and affiliation of the other scientific cooperators:
Koen Venema, Frederik-Jan van Schooten, John Penders and Rob Markus
(Maastricht University), Petra Kuiper and Zsuzsan Proos (NBC), Hetty Busink-van
den Broeck, Ingrid van der Meer, Ruud Timmer (Wageningen University & Research).
Thanks are due to the sponsors for their financial support. The project will be partly
financially supported by the Dutch Topsector AgriFood (TKI 1601P01). Rothamsted
Research receives grant-aided support from the Biotechnology and Biological
Sciences Research Council (BBSRC) of the UK.
References
1. NPD Monitor: www.npd.com
2. Van Gils T, Nijeboer P, IJssennagger CE, et al. Prevalence and characterization of
self-reported gluten sensitivity in The Netherlands. Nutrients 2016; 8: 714; doi:
10.3390/nu8110714.
3. Aziz I, Lewis NR, Hadjivassiliou M, et al. A UK study assessing the population
prevalence of self-reported gluten sensitivity and referral characteristics to
secondary care. Eur J Gatroenterol Hepatol 2014; 26: 33-39.
4. Drossman DA. The functional gastrointestinal disorders and the Rome III process.
Gastroenterology 2006; 130: 1377-1390.
5. Davis W. Wheat Belly. Broadhead Ass 2011; 292 pp.
6. Perlmutter D. Grain Brain. Hodder & Stoughton 2014; 336 pp.
7. Verburg K. De Voedselzandloper. Uitgeverij Bert Bakker 2012; 368 pp.
8. Wu H, Flint AJ, Qibin Q, et al. Association between dietary whole grain intake
and risk of mortality. JAMA Intern Med 2015; 173: 373-384.
9. Huang T, Xu A, Lee A, et al. Consumption of whole grains and cereal fibre and
total and cause-specific mortality: prospective analysis of 367,442 individuals.
BMC Med 2015; 13: 59; doi: 10.1186/s12916-015-0294-7.
10. Aune D, Keum N, Giovannucci E, et al. Whole grain consumption and risk of
cardiovascular disease, cancer, and all cause and cause specific mortality:
systematic review and dose-response meta-analysis of prospective studies. BMJ
2016; 353: i2716; doi: 10.1136/bmj.i2716.
11. Benisi-Kohansal S, Saneei P, Salehi-Marzijarani M, et al. Whole-grain intake and
mortality from all causes, cardiovascular disease, and cancer: a systematic review
and dose-response meta-analysis of prospective cohort studies. Adv Nutr 2016; 7:
1052-1065; doi: 10.3945/an.115.011635.
12. Chen G-C, Tong X, Xu J-Y, et al. Whole-grain intake and total, cardiovascular,
and cancer mortality: a systematic review and meta-analysis of prospective
studies. Am J Clin Nutr 2016; 104: 164-172.
13. Canavan C, West J, Card T. The epidemiology of irritable bowel syndrome. Clin
Epidemiol 2014; 6: 71-80.
14. Uhde M, Ajamian M, Caio G, et al. Intestinal cell damage and systemic immune
activation in individuals reporting sensitivity to wheat in the absence of coeliac
disease. Gut 2016; 65: 1930-1937; doi:10.1136/gutjnl-2016-311964.
15. Caio G, Volta U, Tovoli U, et al. Effect of gluten free diet on immune response to
gliadin in patients with non-celiac gluten sensitivity. BMC Gastroenterology 2014;
14: 2-7.
16. El-Salhy M, Hatlebakk JG, Gilja OH et al. The relation between celiac disease,
nonceliac gluten sensitivity and irritable bowel syndrome. Nutrition Journal 2015;
14: 92.
17. Henström M, D’Amato M. Genetics of irritable bowel syndrome. Mol Cell Pediatr
2016; 3: 7; doi: 10.1186/s40348-016-0038-6.
18. Sofi F, Whittaker A, Cesari F, et al. Characterization of Khorasan wheat (Kamut)
and impact of a replacement diet on cardiovascular risk factors: cross-over dietary
intervention study. Eur J Clin Nutr 2013; 67: 190-195.
19. Sofi F, Whittaker A, Gori AM, et al. Effect of Triticum turgidum subsp.
Turanicum wheat on irritable bowel syndrome: a double-blinded randomised
dietary intervention trial. Brit J Nutr 2014; 111: 1992-1999.
20. Schuppan D, Zevallos V. Wheat amylase trypsin inhibitors as nutritional activators
of innate immunity. Dig Dis 2015; 33: 260-263.
21. Zevallos VF, Raker V, Tenzer S, et al. Nutritional wheat amylase-trypsin
inhibitors promote intestinal inflammation via activation of myeloid cells.
Gastroenterology 2017; DOI: 10.1053/j.gastro.2016.12.006.
22. Mujagic Z, Tigchelaar EF, Zhernakova A, et al. A novel biomarker panel for
irritable bowel syndrome and the application in the general population. Sci Rep
2016; 6: doi: 10.1038/srep26420.
23. Costabile A, Santarelli S, Claus S, et al. Effect of breadmaking process on in vitro
gut microbiota parameters in irritable bowel syndrome PloS One 2014; 9:
e111225. doi:10.1371/journal.pone.0111225.
5. Clinical research reports
Oats in the diet of children with coeliac disease: a
double-blind, randomised, placebo-controlled
multicenter study
Tiziana Galeazzi1, Simona Gatti1 Nicole Caporelli1, Elena Lionetti1, Ruggero
Francavilla2, Maria Barbato3, Paola Roggero4, Basilio Malamisura5, Giuseppe
Iacono6, Andrea Budelli7, Rosaria Gesuita8, Carlo Catassi1
1 Department of Pediatrics, Università Politecnica delle Marche, Ancona, Italy
2 Interdisciplinary Department of Medicine, University of Bari, Bari, Italy
3 Department of Pediatrics, Sapienza University, Rome, Italy
4 Neonatal Intensive Care Unit, Department of Clinical Sciences and Community
Health, IRCCS Ca’ Granda Ospedale Maggiore Policlinico, University of Milan,
Milan, Italy
5 Department of Pediatrics, S. Maria dell’Olmo Hospital, Cava de’ Tirreni, Salerno,
Italy
6 Pediatric Gastroenterology Unit, “G. Di Cristina” Children Hospital, Palermo, Italy
7 R&D Heinz Italia S.p.a., Latina, Italy
8 Department of Epidemiology, Biostatistics, Università Politecnica delle Marche,
Ancona, Italy
Introduction
Coeliac disease (CD) is an inherited autoimmune condition triggered in genetically
susceptible individuals by amino acid sequences within the prolamin fraction of
ingested wheat (gliadins), barley (hordeins) and rye (secaline).
At present, the only treatment for this condition is the lifelong complete withdrawal of
gluten from the diet [1].
Strict adherence to a gluten-free diet (GFD) is required to control the symptoms of CD
and to prevent the autoimmune and neoplastic complications associated with this
condition [2].
However, full compliance with a GFD heavily affects dietary choice and the quality of
life. Although the quality of gluten-free food has significantly improved in the last
decades, some problems still remain partially unresolved, in particular the lower
technological performance of gluten-free cereals [3].
On this basis, the inclusion of oats in the GFD could be of great value. Oats are a good
source of fibres and, in particular, of vitamins and minerals, and beta-glucans, which
are healthy compounds that reduce LDL-cholesterol and the glycemic index of
foodstuff [4,5]. Moreover, the inclusion of oats unquestionably improves the nutritional value and increases the palatability of gluten-free products, while
expanding food choices and ultimately improving the quality of life for people with
CD [6].
Although oats are included among the gluten-free ingredients (gluten content does not
exceed 20 parts per million -ppm-) by European Commission Regulation No. 41/2009
[7] the safety of oats in CD is still a matter of debate.
Some clinical trials have concluded that oats are well tolerated by CD patients on a
GFD, but early studies found that some patients consuming oats as part of a GFD
suffered an intestinal inflammation similar to that in untreated coeliac patients [8].
Previous studies were limited by small sample sizes or short follow-up periods and to
the best of our knowledge there has been only one randomised placebo-controlled
clinical trial.
Moreover, oats is not a staple food in the diet of Mediterranean populations. This is
probably the main reason why an oats “resurrection” in the GFD has not raised
immediate interest in Southern European countries.
Therefore, we aimed to evaluate in a large randomised, double-blind, cross sectional
placebo-controlled multicenter clinical trial the clinical, serological and mucosal safety
and the acceptance of gluten-free oat-based products from selected oat varieties in the
diet of Italian children with CD.
Clinical monitoring during the study was based on: a) score of intestinal symptoms; b)
serological CD markers (TG-IgA, AGA-IgA, antiavenin) c) mucosal parameters
integrity monitored by double sugars intestinal permeability test (IPT).
Material and methods
306 children aged 4-14 years with biopsy-proven diagnosis of CD, on a GFD for at
least 2 years, were recruited at 8 Pediatric Gastroenterology centres in Italy (Ancona,
Bari, Bolzano, Catania, Monza, Palermo, Roma, Cava de’Tirreni) between 2008 and
2012. Patients who (1) have chronic conditions, including type 1 diabetes or
inflammatory bowel disease, or (2) did not adhere to the GFD as demonstrated by
elevation of serological markers at enrollment, were excluded.
Children were assigned on the basis of a stratified randomization to one of two groups:
those assigned to group A received 6 months of diet A, 3 months of standard GFD as a
wash out period, and 6 months of diet B; those assigned to group B received 6 months
of diet B, 3 months of standard GFD as a wash out period, and 6 months of diet A
(Fig. 1).
Figure 1. Study cross-over design
A and B diets included gluten-free products (flour, pasta, biscuits, cakes and crispy
toast) with either purified oats or placebo, that were identical in form, and colour, and
were both provided by a company leader in gluten-free production in Italy.
The minimum oat intake required (calculated as 1 g/kg/day) was 15 g/day for children
aged 3-6 years, 25 g/day for children aged 7-10 years and 40 g/day for children aged
11-16 years. The oat varieties used were specially grown, milled and packaged to
avoid any cross-contamination with gluten-containing cereals or food.
Gluten contamination of used oats was measured by Ridascreen® ELISA test.
Clinical [Body Mass Index (BMI), class of BMI, Gastrointestinal Symptoms Rating
Scale (GSRS) score], serological [IgA anti-transglutaminase antibodies, IgG antideamidated
gliadin peptides (AGA) and IgA anti-avenin] and intestinal permeability
test (IPT) data were measured at basal (B1, recruitment) and after six months of diet A
or B in the first period (T6), after three months of wash-out at the beginning of the
second period as a second basal point (B2) and after six months of diet A or B (T15).
At each time point of follow-up the daily intake of oat was assessed by means of a
three-day diary and symptoms and/or side effects related to the ingestion of the
products under investigation was promptly recorded.
Statistical analysis
Sample size was estimated using intestinal permeability test (IPT) as primary response
variable and considering a clinical difference between the two diets of 0.01 as
minimum.
Since the resulting data was not normally distributed, a non parametric approach was
used for all statistical analyses. For descriptive purposes, absolute variations in clinical
and anthropometric variables between T6 and B1 and T15 and B2, respectively in the
first and in the second period of treatment, were calculated and graphically represented
by boxplots.
Ninety-five percent confidence intervals (95% CI) for median values were calculated
and comparisons between diets groups in each treatment period were performed using
Wilcoxon rank sum test. A positive variation indicated an increase in the variable, that
was considered statistically significant when 95% CI did not contain zero value.
First and second carry-over effect (θ, λ) and direct treatment effect (τ) were evaluated
by a non-parametric statistical approach using medians as summary statistic.
Confidence intervals for each effect were estimated using a probability of 0.90 for the
first two (θ, λ) and 0.95 for τ. A probability of 0.05 was chosen to assess the statistical
significance.
Results and discussion
After the exclusion of 129 patients who dropped out, the analysed cohort included 177
children (79 in group A and 98 in group B). There were 124 girls (70%) and the
median age of the cohort was 8.9 years (range 6.9 to 11.2). Table 1 shows the patients’ main anthropometric and clinical characteristics at the first basal. No significant differences were found between the two groups, so it can be confirmed that
randomization worked well.
No significant differences were found between the two groups in the two treatment
periods both for clinical parameters and serological parameters, neither for the
mucosal parameteres as reported in Fig. 2, 3 and 4, respectively.
Table 1. Subjects' anthropometric and clinical characteristics at basal according to
treatment groups. M=males, F=females, IQR=interquartile range.
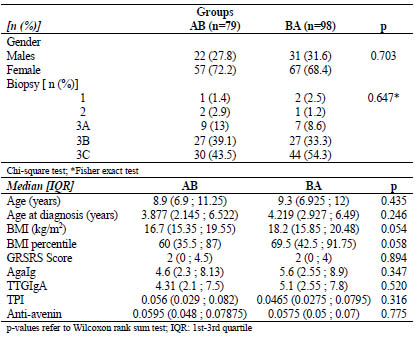
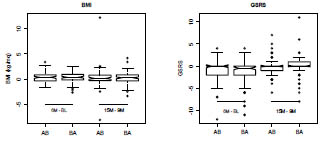
Figure 2. Absolute variation in the two treatment periods according to groups in the
clinical parameters BMI, BMI class and GSRS
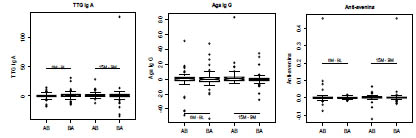
Figure 3. Absolute variation in the two treatment periods according to groups in the
serological parameters TTG, IgA, Aga IgG, anti-avenin)
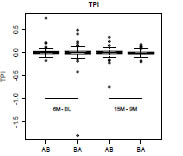
Figure 4. Absolute variation in the two treatment periods according to groups in the
mucosal parameters (intestinal permeability, TPI)
Table 2. Differences in clinical, serological and mucosal parameters during the two
diet periods.
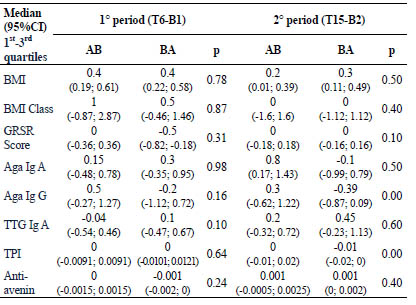
Table 3. Results of crossover analysis between the two study groups.
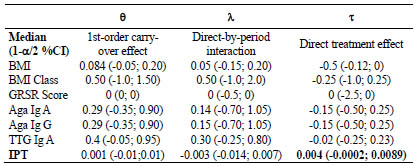
Table 3 shows the results of crossover analysis; differences between the two groups in
the variables of interest were summarised using medians and the first order carry-over
effect, direct-by-period interaction, and a direct treatment effect according to the diet
sequences AB, BA was evaluated by means of confidence intervals. A positive sign in the direct treatment effect estimates indicated that treatment B was associated with the
higher average of the variable.
Differences in treatment carry-over at the time of the second baseline measurements
(θ) and differences in treatment carry-over at the time of the second treatment
measurement (λ) and direct treatment effect (τ) were found not statistically significant
for all clinical, serological and mucosal variables studied. The upper limit of the 95%
confidence interval of IPT direct treatment effect was found lower than the highest
difference considered clinically relevant (0.01).
These data confirm that this clinical trial is a study of non-inferiority, so an oatsenriched
diet did not cause any modifications in coeliac children. There is no
difference in treatment A with respect to treatment B.
Conclusions
In a large group of CD children, we found that the prolonged daily intake of a
considerable amount of pure oats did not cause any significant change in terms of
clinical symptoms, serological parameters and intestinal permeability.
Coeliac disease children can safely add pure oats to their GFD. The inclusion of oats in
the GFD would be beneficial, as they provide a good source of fibres, have a higher
satiety value than other cereals, add texture and flavor to baked goods and could
increase compliance with a GFD by providing patients with more alternatives.
Disclosure of interest: All authors have conflict of interest with Heinz Company s.r.l.
References
1. Rashtak S, Murray JA. Review article: coeliac disease, new approaches to therapy.
Aliment Pharmacol Ther 2012; 35: 768-781.
2. Silano M, Volta U, Mecchia AM, et al. Delayed diagnosis of coeliac disease
increases cancer risk. BMC Gastroenterol 2007; 9: 7.
3. De la Hera E, Talegon M, Caballero P, et al. Influence of maize flour particle size
on gluten-free breadmaking. J Sci Food Agric 2013; 93: 924-932.
4. Wolover TM, Tosh SM, Gibbs AL, et al. Physicochemical properties of oat betaglucan
influence its ability to reduce serum LDL cholesterol in humans: a
randomized clinical trial. Am J Clin Nutr 2010; 92: 723-732.
5. Granfeldt Y, Nyberg L, Bjorck I. Muesli with 4 gram oat beta-glucans lower
glucose and insulin responses after a bread meal in healthy subjects. Eur J Clin
Nutr 2008; 62: 600-607.
6. Rashid M, Butzner D, Burrows V, et al. Consumption of pure oats by individuals
with celiac disease: a position statement by the Canadian Celiac Association. Can
J Gastroenterol 2007; 21: 649-651.
7. Commission Regulation n.41/2009 concerning the composition and labelling of
foodstuff suitable for people intolerant to gluten. Official Journal of the European
Union dated 21.01.2009, L16/3.
8. Silano M, Pozo EP, Uberti F, et al. Diversity of oat varieties in eliciting the early
inflammatory events in celiac disease. Eur J Nutr 2014; 53: 1177-1186.
A study of morphological and immunological
responses to a 14 day gluten challenge in adults with
treated coeliac disease
Vikas K. Sarna1,2, Gry I. Skodje2,3, Henrik M. Reims4, Louise F. Risnes1,5, Shiva D.
Koirala1,5, Ludvig M. Sollid1,2,5, Knut E.A. Lundin2,5,6
1 Department of Immunology and Transfusion Medicine, Oslo University Hospital,
Norway
2 K G Jebsen Coeliac Disease Research Centre, University of Oslo, Norway
3 Division of Cancer Medicine, Oslo University Hospital, Norway
4 Department of Pathology, Oslo University Hospital, Norway
5 Centre for Immune Regulation, University of Oslo, Norway
6 Department of Gastroenterology, Oslo University Hospital, Norway
Introduction
Coeliac disease (CD) is a gluten-induced enteropathy [1]. The treatment is to exclude
gluten completely from the diet, whereupon the derangement of the gut and the serum
antibodies normalise. The clear association to distinct HLA-types has also shown the
central role of T cells in the pathogenesis of CD [2].
Non-coeliac gluten sensitivity is defined as a gluten-induced disease with similar
symptoms as CD, but without the typical findings in the gut and in the blood [3]. The
prevalence of this condition may be higher than CD in many countries [4], leading to
an increased awareness of gluten-induced disease in the population and a significant
number of people eating a self-prescribed gluten-free diet without a proper diagnosis.
A gluten challenge is performed when the patient has started on a gluten-free diet
without proper diagnosis. According to guidelines [5,6], the recommended dose of
gluten should be at least 3 grams daily, and the duration should be at least 8 weeks, or
2 weeks in the case of strong gluten-related symptoms.
In this study we seek to evaluate different response parameters to a 14 day gluten
challenge in adults with treated coeliac disease.
Materials and methods
We included twenty adults (Fig. 1) with treated coeliac disease for a two week gluten
challenge. The participants were on a gluten-free diet, had normal duodenal biopsies
and normal anti-transglutaminase IgA titers prior to challenge. Challenge was
performed with a low-FODMAP (fermentable oligo-, di-, monosaccharides and
polyols) muesli bar containing 5.7 grams of gluten, taken once daily (Fig. 2). We took
duodenal biopsies at the end of challenge. Blood was drawn at several time points. We used HLA-gliadin-complexes (tetramers) linked to a fluorochrome, along with several
monoclonal antibodies to characterise gluten-specific T cells in blood in a flow
cytometer. Symptoms and quality of life parameters were scored by the use of GSRSIBS,
visual analogue scales and SF-36. Serum samples from the first day of challenge
was collected at baseline, and then every second hour until 6 hours after the first dose
of gluten to look for cytokine changes during the initial exposure. Fecal samples were
collected at several time points during the study to characterise the microbiota and
gluten associated changes herein.
The study is approved by the regional ethical committee of South-East Norway (ref.
2013/1237) and registered at clinicaltrials.gov (NCT02464150).
Figure 2. Timeline showing the course of the study
Results and discussion
The manuscript is in preparation and the data are still unpublished. We therefore
choose not to present the results here.
References
1. Sollid LM, Jabri B. Triggers and drivers of autoimmunity: lessons from coeliac
disease. Nat Rev Immunol 2013; 13(4): 294-302.
2. Lundin KEA, Scott H, Hansen T, et al. Gliadin-specific, HLA-DQ(alpha
1*0501,beta 1*0201) restricted T cells isolated from the small intestinal mucosa of
celiac disease patients. J Exp Med 1993; 178(1): 187-196.
3. Ludvigsson JF, Leffler DA, Bai JC, et al. The Oslo definitions for coeliac disease
and related terms. Gut 2013; 62(1): 43-52.
4. Lundin KEA. Non-celiac gluten sensitivity - why worry? BMC Med 2014; 12: 86.
5. Rubio-Tapia A, Hill ID, Kelly CP, et al. ACG clinical guidelines: diagnosis and
management of celiac disease. Am J Gastroenterol 2013; 108(5): 656-676; quiz
677.
6. Ludvigsson JF, Bai JC, Biagi F, et al. Diagnosis and management of adult coeliac
disease: guidelines from the British Society of Gastroenterology. Gut 2014; 63(8):
1210-1228.
A double-blind placebo-controlled cross-over
challenge with gluten and fructans in individuals with
self-reported gluten sensitivity
Gry I. Skodje1,2, Vikas K. Sarna2,3, Ingunn H. Minelle4, Kjersti L. Rolfsen4, Jane G.
Muir5, Peter R. Gibson5, Marit B. Veierød4,6, Christine Henriksen2,4, Knut EA.
Lundin2,3,7
1. Divison of Cancer Medicine, Oslo University Hospital, Oslo, Norway
2 K.G. Jebsen Coeliac Disease Research Centre, University of Oslo, Norway
3 Faculty of Medicine, University of Oslo, Norway
4 Department of Nutrition, Institute for Basic Medical Sciences, University of Oslo,
Norway
5 Department of Gastroenterology, Monash University and Alfred Hospital,
Melbourne, Victoria, Australia
6 Department of Biostatistics, Oslo Centre for Biostatistics and Epidemiology,
University of Oslo, Norway
7 Department of Gastroenterology, Oslo University Hospital, Oslo, Norway
Introduction
Non-coeliac gluten sensitivity (NCGS) is a term within gluten-related disorders that
has been applied for the condition where persons report symptom relief on a glutenfree
diet in absence of coeliac disease and wheat allergy [1]. An important
characteristic for the group is the experience of symptoms after intake of glutencontaining
cereals, but the role of gluten as symptom inducer is questioned. There are
no reliable diagnostic biomarkers for NCGS, but a standardised blinded placebocontrolled
gluten challenge is proposed to confirm the condition [2].
Results from two earlier double-blinded placebo-controlled studies have given
conflicting results. The first study showed that subjects who received gluten reported
more gastrointestinal symptoms than those who received placebo [3]. The second
study showed no specific or dose-dependent effect of gluten as compared to placebo
after dietary reduction of fermentable, poorly absorbed short-chained carbohydrates
(FODMAP) in subjects who were believed to have NCGS [4]. Subjects in these trials
had coeliac disease excluded by negative HLA-DQ2/8 or normal duodenal biopsy
while on a gluten-containing diet. Several double-blinded placebo-controlled gluten
challenge trials have followed [5-7]. Recent trials suggest that gluten challenge
induces symptom recurrence in only a minority of subjects who meet clinical criteria
for NCGS [6]. Two other Italian trials aimed to discover true NCGS and found
increased symptom score on gluten intake as compared to placebo, but only 5% and
14% were classified as NCGS according to predefined criteria [5,7]. In both studies
gluten was administered as capsules.
All these trials have studied the role of gluten and used carbohydrate depleted gluten
flour as the active challenge vehicle. However, people do not eat pure gluten flour.
They report wheat as the symptom inducer. Considering the composition of wheat,
where gluten co-exists with a substantial amount of FODMAP, in specific fructans,
this component has not been taken into account in any challenge trial of NCGS [8].
The aim of the present study was therefore to investigate the effect of gluten and
fructans separately in self-reported gluten-sensitive persons with muesli bars as
challenge vehicle.
Materials and methods
Study design
We describe a double-blind, placebo-controlled challenge study, where subjects were
randomised to gluten, fructan and placebo challenges in a cross-over design
(ClinicalTrials.gov, NCT02464150). The study took place at Oslo University Hospital,
Rikshospitalet from October 2014 to May 2016.
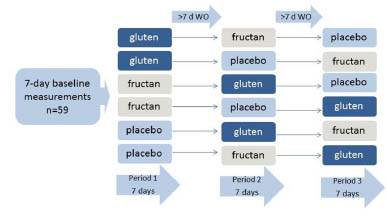
Figure 1. Study design. WO, wash out
Subjects
The study subjects were 59 adults (6 males) aged 18-80 who self-reported gluten
sensitivity, had been strictly adherent to a gluten-free diet (GFD) for at least six
months followed by symptom relief and had no coeliac disease or wheat allergy.
Coeliac disease was considered adequately excluded by negative duodenal biopsy
while on gluten-containing diet or negative genotype HLA-DQ2/8. Wheat allergy was
considered eliminated in case of negative wheat-specific IgE. Exclusion criteria were
pregnancy or lactation, use of immunosuppressive agents, inflammatory bowel diseaseor other comorbidity, substantial infection, long travel distance or allergy to nuts or
sesame seeds.
Subjects were recruited by advertisements on the Oslo University Hospital’s web page,
at the University of Oslo, at the web page of the Norwegian Coeliac Association and
their social media, and by referrals from general practitioners and other hospitals in the
area.
Food challenge protocol
The subjects were randomised to one of three challenges (gluten, fructan or placebo)
for one week, followed by a minimum of one week washout period. They continued
their GFD and were told to keep their diet otherwise consistent with the baseline diet
throughout the study.
The challenge vehicle was a 50 g, 220 kcal low-FODMAP gluten-free muesli bar that
was eaten once a day. The muesli bars were developed and produced by the Monash
University, Melbourne. The fructan bar additionally contained 2.1 g of fructooligosaccharides
and the gluten bar 5.7 g of gluten. The gluten used was commercially
available, carbohydrate depleted wheat gluten (Vital Wheat Gluten, Manildra Group,
Gladesville, New South Wales, Australia) with nutritional content of 75% protein, 9%
carbohydrate of which 4% sugar, 6% fat, 9% water and 1% ash (Cargill analysis). The
muesli bars were balanced in carbohydrates, fiber, fat and protein and had similar
appearance, texture and taste, confirmed by pre-testing in 12 healthy adults where the
gluten bar could not be differentiated from the fructan or placebo bar.
Aim and outcome
The aim was to investigate the effect of gluten and fructans separately in a doubleblind
placebo controlled challenge in individuals with self-reported gluten sensitivity
in absence of coeliac disease. The primary outcome was overall gastrointestinal
symptoms measured by the Gastrointestinal Symptom Rating Scale – irritable bowel
syndrome version (GSRS-IBS). The secondary outcome was daily overall
gastrointestinal symptom score by visual analogue scale (VAS) for abdominal pain,
bloating, passage of wind, nausea, stool dissatisfaction and overall gastrointestinal
symptoms.
Results and discussion
The manuscript is in preparation. Results will therefore not be presented here.
References
1. Ludvigsson JF, Leffler DA, Bai JC, et al. The Oslo definitions for coeliac disease
and related terms. Gut 2013; 62(1): 43-52.
2. Catassi C, Elli L, Bonaz B, et al. Diagnosis of non-celiac gluten sensitivity
(NCGS): The Salerno experts' criteria. Nutrients 2015; 7(6): 4966-4977.
3. Biesiekierski JR, Newnham ED, Irving PM, et al. Gluten causes gastrointestinal
symptoms in subjects without celiac disease: a double-blind randomized placebocontrolled
trial. Am J Gastroenterol 2011; 106(3): 508-514; quiz 515.
4. Biesiekierski JR, Peters SL, Newnham ED, et al. No effects of gluten in patients
with self-reported non-celiac gluten sensitivity after dietary reduction of
fermentable, poorly absorbed, short-chain carbohydrates. Gastroenterology 2013;
145(2): 320-8 e1-3.
5. Di Sabatino A, Volta U, Salvatore C, et al. Small amounts of gluten in subjects
with suspected nonceliac gluten sensitivity: A randomized, double-blind, placebocontrolled,
cross-over trial. Clin Gastroenterol Hepatol 2015; 13: 1604-1612.e3.
6. Zanini B, Baschè R, Ferraresi A, et al. Randomised clinical study: gluten
challenge induces symptom recurrence in only a minority of patients who meet
clinical criteria for non-coeliac gluten sensitivity. Aliment Pharmacol Ther 2015;
42(8): 968-976.
7. Elli L, Tomba C, Branchi F, et al. Evidence for the presence of non-celiac gluten
sensitivity in patients with functional gastrointestinal symptoms: Results from a
multicenter randomized double-blind placebo-controlled gluten challenge.
Nutrients 2016; 8(2): 84.
8. Biesiekierski JR, Rosella O, Rose R, et al. Quantification of fructans, galactooligosacharides
and other short-chain carbohydrates in processed grains and
cereals. J Hum Nutr Diet 2011; 24(2): 154-176.
Natural history and management of potential coeliac
disease
Renata Auricchio, Valentina Discepolo, Roberta Mandile, Maria Maglio, Luigi Greco,
Riccardo Troncone
Department of Medical Translational Sciences & European Laboratory for the
Investigation of Food Induced Diseases, University Federico II, Naples, Italy
Introduction
According to the most recent European Society of Gastroenterology, Hepatology and
Nutrition (ESPGHAN) guidelines, coeliac disease (CD) is considered an immunemediated
systemic disorder elicited by gluten and related prolamins in genetically
susceptible individuals [1]. Even if enteropathy remains the prominent feature of the
disease, it is now widely accepted, that, from a histological point of view, it can range
from complete villous atrophy to minimal mucosal abnormalities. In this context, the
term potential coeliac disease (PCD) is used to define patients with normal or slightly
altered intestinal mucosa (Marsh 0-1), but a positive CD serology. Patients with PCD
may or may not have symptoms and may or may not develop an overt form of CD
over time [2]. The number of patients with a diagnosis of PCD is increased so far
because of the screening of general population and it is now estimated to be at the
considerable number of 1/5-1/10 of the total CD diagnosis [3].
Clinical management of patients with potential coeliac disease
Even if PCD patients do not show clear signs of enteropathy, some of them may
present clinical symptoms. In our experience, abdominal pains and failure to thrive are
the most frequent ones and are found in around 1/3 of symptomatic patients. Other
symptoms are diarrhea (approximately 16% of patients), lack of appetite (13%), low
blood ferritin (8%), vomiting and constipation (5%), anemia and
hypertransaminasemia (3%). Biagi et al. have hypothesised that in PCD the intestinal
mucosa is maintained architecturally normal thanks to an increased enterocytic
proliferation, which, however, will end up in a reduced enterocytic maturity and thus
in a reduced absorptive capacity of the small bowel [4]. There is a general consensus
for this kind of patients to adopt a gluten-free diet (GFD) and to control, during the
follow up, the remission of symptoms.
On the contrary, the management of asymptomatic patients with PCD remains a major
clinical problem. Some have suggested that, because this condition could be the first
step of the disease, all patients should adopt a lifelong GFD. This was supported by the
fact that an undiagnosed case of CD has a four-fold increased risk of mortality for all
causes [5] and that the “metabolic identikit” of PCD patient is similar to the one of CD
patients and differs from controls [6]. Moreover, despite per definition there is no clear villous atrophy in potential coeliac disease, in the last decade an increasing number of
studies have suggested that mild signs of inflammation are often present. In fact, from
an immunohistochemical point of view, 70.8% of PCD patients show increased
numbers of lamina propria CD25+ and/or enhanced expression of ICAM-1 and crypt
HLA-DR [7]. On the other hand, preliminary observations from our group
demonstrated that most of the asymptomatic children with PCD remained healthy on a
gluten-containing diet. During three years of follow up, CD-associated antibodies
fluctuated (32.6%) or even disappeared (14.6%) and, after three years, only 30.8% of
the patients developed villous atrophy [8]. Therefore, it would be an overtreatment to
consider all PCD patients as coeliac. Unfortunately, we still have no good way to
identify which subsets of seropositive patients will develop mucosal damage.
Natural history of potential coeliac disease: a 9 years prospective longitudinal study
The main challenge remains to find criteria that allow to differentiate, among PCD
patients, those who will develop villous atrophy from those who will not. In order to
study the long-term natural history of PCD disease and to explore risk factors
associated with the development of mucosal atrophy, our group performed a 9 year
prospective longitudinal study [9]. 175 asymptomatic children were left on a glutencontaining
diet. Antibodies and clinical symptoms were checked every 6 months, and a
small bowel biopsy was taken every 2 years to evaluate histological,
immunohistochemical, and anti-TG2 deposits. Patients were genotyped for HLA and a
set of non-HLA CD-associated genes. At the end of the follow up, 67% of the patients
still had a normal duodenal architecture (Fig. 1). Monitoring the individual profile of
anti-TG2 antibodies, 43% of patients showed persistently elevated anti-TG2 level,
20% became negative during follow-up, and 37% showed a fluctuant anti-TG2 course
with transiently negative values. Analysing the cumulative incidence of CD in relation
to individual risk factors, we noticed that the anti-TG2 titer at the entry was not
statistically different between those who remained potential and those who progressed
(become atrophic or developed symptoms), but the variation of anti-TG2 correlated to
the final outcome. In fact, none of the negative anti-TG2 patients developed full-blown
disease, whereas among those who developed severe damage, 78.8% had persistently
positive anti-TG2 compared with 43% of those who did not develop the intestinal
damage. Up to date, the detection of intestinal deposits of immunoglobulin A (IgA)
anti-TG2 by immunofluorescence was reported to be the best marker to identify,
among patients with potential CD, those who will eventually develop a glutendependent
enteropathy [8-10]. Moreover, to further explore which variable or risk
factor is likely to differentiate the patients who progress to flat mucosa from those who do not, a multivariate analysis was performed: male sex, slight mucosal inflammation
at time 0 (estimated by the numbers of CD25+ cells and γδ+ lymphocytes) and a
certain genetic profile (genes HLA-DQ2 and polymorphism of IL12A, OLIG3, and
IL2/IL21) may well start to delineate a cohort of individuals who are more prone to
develop the disease.
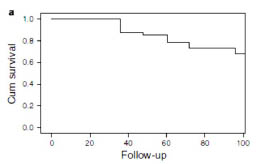
Figure 1. Cumulative survival function in the cohort study. Percentage of cases who
remained potential during follow-up [9]
Conclusion
Potential coeliac disease is a condition more and more frequently diagnosed because of
the screening of the general population, however the clinical management of the
disease remains debated. The presence of symptoms should always induce to prescribe
a gluten-free diet. On the contrary, prescribing a gluten-free diet to all asymptomatic
patients could be an overtreatment, as we demonstrated that only 33% of the patients
will develop mucosal atrophy within 9 years. Unfortunately, there are still no
biomarkers that can allow to differentiate with confidence patients that will develop an
intestinal damage from those who will not. However, preliminary studies suggest that
male sex, the number of CD25+ and γδ+ lymphocytes in the intestinal mucosa at the
time of diagnosis, as well as HLA-DQ dose and some non-HLA polymorphisms may
help to identify a cohort of individuals more prone to develop overt disease.
References
1. Husby S, Koletzko S, Korponay-Szabó IR, et al. European Society for Pediatric
Gastroenterology, Hepatology, and Nutrition Guidelines for the Diagnosis of
Coeliac Disease. J Pediatr Gastroenterol Nutr 2012; 54: 136-160.
2. Ludvigsson JF, Leffler DA, Bai J, et al. The Oslo definitions for coeliac disease
and related terms. 2013; 62(1): 43-52.
3. Volta U, Caio G, Giancola, et al. Features and progression of potential celiac
disease in adults. Clin Gastroenterol Hepatol 2016; 14(5): 686-693.
4. Biagi F, Trotta L, Alfano C, et al. Prevalence and natural history of potential
celiac disease in adult patients. Scand J Gastroenterol 2013; 48(5): 537-542.
5. Rubio-Tapia A, Kyle RA, Kaplan EL. Increased prevalence and mortality in
undiagnosed celiac disease. Gastroenterology 2009; 137: 88-93.
6. Bernini P, Bertini I, Calabrò A, et al. Are patients with potential celiac disease
really potential? The answer of metabonomics. J Proteome Res 2011; 10(2): 714-
721.
7. Paparo F, Petrone E, Tosco A, et al. Clinical, HLA, and small bowel
immunohistochemical features of children with positive serum antiendomysium
antibodies and architecturally normal small intestinal mucosa. J Gastroenterol
2005; 100(10): 2294-2298.
8. Tosco A, Salvati VM, Auricchio R, et al. Natural history of potential celiac
disease in children. Clin Gastroenterol Hepatol 2011; 9: 320-325.
9. Auricchio R, Tosco A, Piccolo E, et al. Potential celiac children: 9-year follow-up
on a gluten-containing diet. Am J Gastroenterol 2014; 109(6): 913-921.
10. Korponay-Szabó IR, Halttunen T, Szalai Z, et al. In vivo targeting of intestinal and
extraintestinal transglutaminase 2 by celiac autoantibodies. Gut 2004; 53: 641-648.
Association between IL-33/ST2 axis and active
coeliac disease
Federico Perez1, David Diaz Jimenez2, Carolina N. Ruera1, Marjorie de la Fuente2,
Glauben Landskron2, Agustina Redondo4, Luciana Guzman3, Marcela Hermoso2,
Fernando Chirdo1
1.Instituto de Estudios Inmunológicos y Fisiopatológicos, Facultad de Ciencias
Exactas, Universidad Nacional de la Plata, Argentina
2 Instituto de Ciencias Biomédicas, Universidad de Chile, Santiago, Chile
3 Servicio de Gastroenterología Hospital de Niños "Sor María Ludovica" de La Plata,
Argentina
4 Servicio de Gastroenterología HIGA San Martin, La Plata, Argentina
Introduction
IL-33, a member of the IL-1 family, is mainly expressed by epithelial, endothelial and
mesenchymal cells [1]. In resting cells, it is located principally in the nucleus, where it
can regulate the expression of certain genes by its interaction with different
transcriptional factors. It has been proven that this protein can be released from the
nucleus into the cytoplasm and then to the extracellular space by different kinds of
stimuli inducing a strong inflammatory response through its receptor, ST2. The IL-33
receptor is a heterodimeric complex made up of two different proteins ST2L, which
actually binds IL-33 and an accessory protein known as IL-1RAc. This receptor is
expressed by different cells: lymphocytes, mast cells, NK cells, ILC2, and endothelial,
epithelial and mesenchymal cells. The ST2L protein has a splice variant, named as
soluble ST2 (sST2) which can be released into the extracellular space where it works
as decoy factor for IL-33 [2-3].
Since the discovery of IL-33 many different functions have been linked to this
cytokine. Firstly, it was recognised for its capacity to promote Th2 responses [4]. More
recently, it has been shown that IL-33 may also promote Th1 functions [5]. It is clear
that IL-33 acts not only as a cytokine, but also as alarm signal, stimulating many proinflammatory
responses [6]. Particularly, necrotic cells, but not apoptotic ones, release
an active form of IL-33 with potent biological effects [2]. Based on the functional
properties of this molecule, we aimed to investigate whether IL-33 may play a role in
coeliac disease (CD) pathogenesis. It has been recently observed that CD patients
present increased levels of IL-33 [2]. In the present work, we evaluate the expression
of IL-33 and its receptor in human intestinal mucosa. These are the initial studies in
order to link the biology of IL-33 and the mechanisms of CD pathogenesis.
Materials and methods
Blood samples from a total of 40 untreated CD patients and 39 healthy controls were
evaluated in this study. Duodenal biopsies from 9 untreated CD patients and 9 healthy
controls were used to perform immunofluorescence assays. All the samples were taken
during the routine protocol for CD diagnosis in the Gastroenterology Units of Hospital
Sor María Ludovica (paediatric patients) and Hospital San Martin (adult patients). The
study was approved by the ethics committee of both institutions.
For immunofluorescence analysis, 4 μm sections of duodenal paraffin-embedded
tissues were used. Antigen retrieval was performed by heat treatment in citrate buffer,
and stained with commercial antibodies from R&D systems (AF3625 for IL-33 and
AF523 for ST2). Nuclei were stained with DAPI.
Images were obtained using a SP5 Leica confocal microscope. The cellular density
was calculated as the number of positive cells for each marker in a predefined zone.
Levels of ST2 and IL-33 in serum samples were determined by commercial ELISA
kits (R&D System, cat. DY523 for ST2, and ab119547 for IL-33).
Comparison of the expression levels for IL-33 and ST2 in serum and positive cell
numbers in lamina propria between control subjects and active CD patients was
performed using unpaired T-tests with 95% of confidence.
Results and discussion
First, we determined the concentration of circulating IL-33 and the soluble form of
ST2 (sST2) in untreated CD patients and healthy controls by commercial ELISA.
Increased levels of sST2 in sera of untreated CD patients were observed. The mean
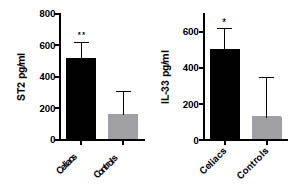
Figure 1. Concentration of IL-33 and its decoy receptor (sST2) in serum samples of
untreated CD patients (n=37 for sST2 and 21 for IL-33) and healthy controls (n=39
for sST2 and 9 for IL-33). Plots show the mean value plus SEM bars of the
concentration of IL-33 and sST2 (pg/ml). (IL-33, *p<0.01, sST2: ** p<0.005)
value of sST2 was 264,1 pg/ml for untreated CD patients while 96.5 pg/ml was
observed for healthy controls (Fig. 1). Untreated CD patients presented also increased
levels of IL-33. The mean value for CD patients was 277.1 pg/ml and 15.13 pg/ml for
healthy controls.
Next, we aimed to evaluate the expression of IL-33 and ST2 in the intestinal mucosa.
To this end, we evaluated sections of duodenal biopsies from untreated CD patients
and healthy controls by immunofluorescence. The immunofluorescence images
showed a higher number of IL-33 and ST2 expressing cells in the lamina propria of
active CD patients than in controls. The epithelium showed a faint staining. However,
in some CD patients some cells from the epithelial compartment showed a strong
staining for both proteins.
The ST2+ cells were located close to the epithelium. IL-33+ cells did not appear
randomly distributed. On the contrary, they seem to be organised or associated with
some structures resembling blood vessels.
In order to quantify the number of IL-33+ or ST2+ cells located in the lamina propria,
we performed manual counting of each group of cells. Cells were counted per m2 of
lamina propria to obtain the density of positive cells. Higher numbers of IL-33+ and
ST2+ cells were observed in duodenal lamina propria from untreated CD patients (Fig.
2).
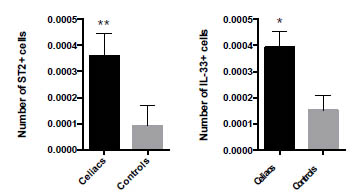
Figure 2. Increased numbers of IL-33+ cells and ST2+ cells in duodenal lamina
propria from untreated CD patients. Counting of IL-33+ cells and ST2+ cells in
immunofluorescence images of predefined areas of duodenal lamina propria of
untreated CD patients (n=9) and healthy controls (n=9). Plots show the mean value
plus SEM bars of the number of IL-33+ cells and ST2+ cells per m2 of lamina propria,
(ST2 ** p<0.05, IL-33 * p<0.1)
Since CD is a very well-known Th1 driven enteropathy [8], the high expression of IL-
33 in duodenal mucosa and in peripheral blood of untreated CD patients is intriguing.
Recent works have demonstrated the broad spectrum of effects of IL-33 in different
pathologies. Yang et al. proved that only cytotoxic T cells in a context Th1 or Th17
express ST2 [9]. These authors also confirmed that IL-33 synergises with IL-12 to
induce effector cells. In addition, other studies confirmed that IL-33 and ST2
expression was necessary to mount a cytotoxic response against some virus infections
and tumours, highlighting the role of the IL-33/ST2 axis in the induction of potent
cytotoxic T cells [10-11]. Bourgeois et al. showed that IL-33 directly interacts with
iNKT and NK cells to induce IFN-γ production [12]. On the other hand, IL-33
enhanced Th1/Th17 responses in some mouse models [13-14]. Therefore, we
hypothesise that the IL-33/ST2 axis may have a role in CD pathogenesis, probably
expanding the inflammatory process, and promoting a Th1 and cytotoxic response.
Further work is in progress to investigate the signal involved in the up-regulation of
this factor and also the consequences of its systemic release.
Conclusion
In this study, we found higher levels of IL-33 and sST2 in serum of untreated CD
patients compared with healthy controls, together with an increase in the number of
IL-33+ and ST2+ cells in duodenal lamina propria of untreated CD patients.
Since IL-33 is mainly located in the nucleus of different cells, upregulation of IL-33
expression points to its role as alarm signal.
Though the test used to evaluate serum IL-33 levels does not discriminate active and
inactive form, it is likely that part of the circulating IL-33, released from the intestinal
mucosa, may reach distant tissues initiating a tissue damage process. IL-33 together
with other cytokines, such as IFN- and IL-15 may also be part of the inflammatory
mediators which link CD to other inflammatory/autoimmune diseases such as type I
diabetes and rheumatoid arthritis.
References
1. Moussion C, Ortega N, Girard JP. The IL-1-like cytokine IL-33 is constitutively
expressed in the nucleus of endothelial cells and epithelial cells in vivo: A novel “Alarmin”? PLoS One 2008; 3(10): 1-8.
2. Martin MU. Special aspects of interleukin-33 and the IL-33 receptor complex.
Semin Immunol Elsevier Ltd 2013; 25(6): 449-457.
3. Martin NT, Martin MU. Interleukin 33 is a guardian of barriers and a local
alarmin. Nat Immunol Nature Publishing Group 2016; 17(2): 122-131.
4. Schmitz J, Owyang A, McClanahan TK, et al. IL-33, an interleukin-1-like
cytokine that signals via the IL-1 receptor-related protein ST2 and induces T
helper type 2-associated cytokines. Immunity 2005; 23(5): 479-490.
5. Baumann C, Bonilla WV., Hegazy AN, et al. T-bet– and STAT4–dependent IL-33
receptor expression directly promotes antiviral Th1 cell responses. Proc Natl Acad
Sci 2015 Mar 31; 112(13): 4056-4061.
6. Haraldsen G, Balogh J, Sponheim J, et al. Interleukin-33 cytokine of dual function
or novel alarmin? Trends Immunol 2009; 30(5): 227-233.
7. López-Casado M, Lorite P, Palomeque T, et al. Potential role of the IL-33/ST2
axis in celiac disease. Cell Mol Immunol 2017; 14: 285-292.
8. Nilsen EM, Jahnsen FL, Sollid LM, et al. Gluten induces an intestinal cytokine
response strongly dominated by interferon gamma in patients with celiac disease.
Gastroenterology 1998; 115(3): 551-563.
9. Yang Q, Li G, Zhu Y, et al. IL-33 synergizes with TCR and IL-12 signaling to
promote the effector function of CD8+ T cells. Eur J Immunol 2011; 41(11):
3351-3360.
10. Bonilla WV, Frohlich A, Johnson S, et al. The alarmin interleukin-33 drives
protective antiviral CD8+ T cell responses. Science 2012; 335(6071): 984-989.
11. Gao X, Wang X, Li G, et al. Tumoral expression of IL-33 inhibits tumor growth
and modifies the tumor microenvironment through CD8+ T and NK cells. J
Immunol 2014; 194(1): 438-445.
12. Bourgeois E, Van LP, Samson M, et al. The pro-Th2 cytokine IL-33 directly
interacts with invariant NKT and NK cells to induce IFN-γ production. Eur J
Immunol 2009; 39(4): 1046-1055.
13. Palmer G, Seemayer CA, Viatte S, et al. Inhibition of interleukin-33 signaling
attenuates the severity of experimental arthritis. Arthritis Rheum 2009; 60(3): 738-
749.
14. Xu D, Jiang H-R, Fraser AR, et al. IL-33 exacerbates antigen-induced arthritis by
activating mast cells. Proc Natl Acad Sci U S A. 2008; 105(31): 10913-10918.
Abrogation of coeliac immunogenicity of gluten
peptides by amino acid point substitutions
Nika Japelj1, Beatriz Côrtez-Real1, Tanja Šuligoj1, Wei Zhang2, Joachim Messing2,
Uma Selvarajah1, Paul J. Ciclitira1
1 Department of Gastroenterology, Kings College, St Thomas Hospital, London,
United Kingdom
2 Rutgers University, Waksman Institute of Microbiology, New Jersey, USA
Introduction
The only generally accepted treatment of coeliac disease (CD) is a life-long gluten-free
diet. Wheat gluten proteins include gliadin, low- (LMWG) and high-molecular-weight
glutenins (HMWG), all three of which have shown to be CD-toxic [1,2,3]. A glutenfree
diet significantly reduces the quality of life for affected individuals such that new
approaches are sought. We have identified naturally existing variants of gliadins and
glutenins that might be less immunotoxic in CD [4].
Aims
We sought to test selected variants of α-gliadin peptides in CD T-cell proliferation
assays with gluten-sensitive T-cell lines that had been generated from duodenal
biopsies from individuals with CD. This would allow us to evaluate their CD-toxic
immunogenicity. We sought to identify peptides with lower CD-toxic immunogenicity
as a prelude to providing the basis of new dietary strategies as part of a gluten-free diet
for individuals with CD.
Materials and methods
We generated gluten-specific polyclonal T-cell lines from duodenal biopsies taken
from individuals with CD (n=11) as previously described [5] although not all the Tcell
lines were used in every study. The candidate peptides, as shown in Fig. 1, were
tested in proliferation assays using radioactive labelled thymidine to measure T-cell
proliferation. A stimulation index >2 was considered positive. We tested five α-gliadin
peptides synthesised as 16mers. The results of proliferation T-cell assays with medium
alone, PT digested gluten, peptides 1, 3, 5, 6, and 9 are presented in Tab. 1. The set of
tested peptides harbored the overlapping T-cell epitopes DQ2.5-glia-α-1a and DQ2.5-
glia-α-2, and naturally occurring variants that differed in a few amino acids (shown in
Fig. 1).

Table 1: Results of 11 proliferation assays: 11 gluten-sensitive T-cell lines were tested with medium only, peptic tryptic
digested gluten and peptides 1, 3, 5, 6 and 9. Stimulation indices for individual antigens are marked in bold and
corresponding arithmetic mean ± standard deviation in brackets. Note that SI greater than 2 were considered as
positive.
Results and discussion
Approximately half of gluten-specific cell lines (5:10) recognise immune-dominant
peptide 1 (QLQPFPQPQLPYPQPQ) or its deamidated counterpart
(QLQPFPQPELPYPQPQ, peptide 3). Stimulation indices vary from 0.7-14.76. That
confirms different sensitivity of T cells obtained from different patients to particular
epitopes. Notably, indices increase when peptide 1 is deamidated (Fig. 2).
Peptide 3’s point substituted variant (QLQPFPQPELSYPQPE, Peptide 5) triggered
positive T-cell responses in 2:6 CD gluten-sensitive T-cell lines, the results of which
are shown in Fig. 3. Peptides with two substitutions (QLQPFQPKLSYPQPE, Peptide
6) or three amino acid substitutions (QLQPFPKPKPKLPYPKPQ, Peptide 9) did not
stimulate any tested gluten-sensitive T-cell lines (n=8 and n=5, respectively). The
above results indicate the importance of both deamidated glutamic acid (at position 65,
Fig. 1) and proline (at position 67, Fig. 1) in triggering coeliac-specific reactions.
Conclusion
We have shown that introduction of two selected amino acid substitutions in α-gliadin
peptides abrogates responses of CD gluten-sensitive T cells to these peptides. We
suggest that these peptides will need additional assessment using CD small intestinal
biopsy organ culture and in vivo testing to confirm their lack of CD toxicity.
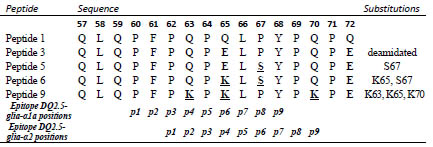
Figure 1. Amino acid sequences for α-gliadin peptides and substituted variants used in
the study. Letters in bold indicate amino acid substitutions within the
immunodominant p57-72 α-gliadin peptide. The bottom part of the table shows
epitopes within this peptide and the position of their binding into the HLA-DQ2 groove
(p1-9)

Figure 2. Proliferative response of T-cell lines to immunodominant peptide 1 (epitope
DQ2.5-glia-α1a positions and DQ2.5-glia-α2) and to deamidated counterpart
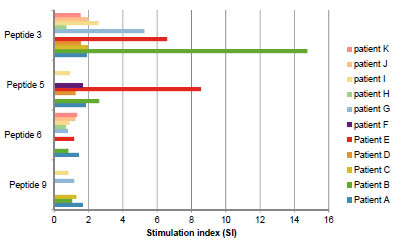
Figure 3. Proliferative response of T-cell lines to different α-gliadin peptides; peptide
3, peptide 5, peptide 6 and peptide 9. With every further peptide, one amino acid
substitution is introduced. With 2 amino acid substitutions, T-cell response is
completely abrogated
References
1. Ciclitira PJ, Evans DJ, Fagg NL, et al. Clinical testing of gliadin fraction in
coeliac patients. Clin Sci 1984; 66: 357-364.
2. Vader W, Kooy Y, van Veelen P, et al. The gluten response in children with
coeliac disease is directed towards multiple gliadin and glutenin peptides.
Gastroenterology 2002; 122: 1729-1737.
3. Dewar DH, Amato M, Ellis HJ, et al. The toxicity of high molecular weight
glutenin subunits of wheat to patients with coeliac disease. Eur J Gastroenterol
2006; 18: 493-491.
4. Zhang W, Ciclitira PJ, Messing J. PacBio sequencing of gene families – A case
study with wheat gluten genes. Gene 2014; 533:(2) 541-546.
5. Molberg O, McAdams S, Lundin KE, et al. T cells from coeliac disease lesions
recognize gliadin epitopes deamidated in situ by endogenous tissue
transglutaminase. Eur J Immunol 2001; 31: 1317-1323.
Estimation of (sero) prevalence of coeliac disease in
children and adolescents in the LIFE Child study
cohort
Johannes Wolf1, Norman Haendel2, Anne Jurkutat3, Carl Elias Kutzner1, Gunter
Flemming2, Wieland Kiess2,3, Andreas Hiemisch2,3, Antje Körner2,3, Wolfgang
Schlumberger4, Joachim Thiery1,3, Thomas Mothes1
1 Institute of Laboratory Medicine, Clinical Chemistry and Molecular Diagnostics,
Medical Faculty of the University and University Hospital, Leipzig, Germany
2 Department of Women and Child Health, Hospital for Children and Adolescents and
Centre for Paediatric Research (CPL), University of Leipzig, Germany
3 LIFE Leipzig Research Centre for Civilization Diseases, University of Leipzig,
Germany
4 EUROIMMUN Medizinische Labordiagnostika AG, Lübeck/Dassow, Germany
Introduction
The prevalence of coeliac disease (CD) in population-representative cohorts is
reported between 0.18 and 2.38% [1] depending on the size and mean age of the
screened population as well as on the applied serological tests.
Currently, four studies are available estimating the frequency of CD in Germany [1-4]
which screened in different cohorts (paediatric and/or adult cohorts) and applied
varying screening strategies. The prevalence of biopsy-proven CD was found between
0.18 and 0.37% [1,2]. This seems to be underestimated considering the frequency of
CD in other European countries [5]. Contrary, seroprevalences in randomly selected
cohorts were found between 0.8 and 1.35% [3,4]. This suggests a higher frequency of
CD in Germany but might be due to a certain low degree of unspecificity of the
applied antibody tests. Otherwise, a recent screening [6] demonstrated that 98% of the
participants with IgA anti-tissue transglutaminase (IgA-aTTG) values ≥10 times of
upper limit (ULN) exhibit mucosal impairment compatible with CD (≥ Marsh 3A).
The primary objective of our ongoing study was to estimate (sero)prevalences in a
paediatric randomly selected German cohort by applying additionall confirmatory
antibody tests and HLA genotyping. Furthermore, we asked the question if antibody
test results alone can predict the actual prevalence of CD in a paediatric cohort.
Materials and methods
Study population
The Leipzig Research Centre for Civilization Diseases (LIFE) Child study has been
designed to understand how and through which mechanisms and mediators genetic, metabolic and environmental factors influence health and development in children and
adolescents [7]. LIFE Child is a prospective, longitudinal population-based cohort
study of urban children from fetal life until adulthood. The study focuses on
monitoring of normal growth, development and health non-communicable diseases
such as childhood obesity, atopy and mental health problems. Families were randomly
selected and invited by the residents’ registration office. Between 2011 and 2015, 3080
children and adolescents were included, of whom 2363 participants were enrolled for
our CD screening due to the inclusion criteria (age between 1 and 18 years, at least
two serum aliquots of the first visit with blood withdrawal available).
Study procedure
The study procedure is shown in Fig. 1. We performed a four-step CD screening. The
first step comprised determination of IgA-aTTG and IgG-antibodies against
deamidated gliadins (IgG-aDGL) in sera (T0) using tests of EUROIMMUN AG
(Lübeck, Germany). Afterwards, samples with positive results for IgA-aTTG
(including also samples with IgA-aTTG values between 0.5 and 1 x ULN) and for

Figure 1. Study procedure. CD coeliac disease, EmA endomysium antibodies, GFD
gluten-free diet, IgA-aTTG IgA anti-tissue transglutaminase, IgG-aDGL IgGantibodies
against deamidated gliadins, JIRA juvenile idiopathic rheumatoid arthritis,
T1 follow-up in LIFE Child, T1DM diabetes mellitus type I, T2 personal interview,
ULN upper limit of normal
IgG-aDGL or samples of patients with CD-associated disorders as well as known CD
were tested for endomysium antibodies (EmA) and HLA-DQ2 (HLA-DQ2.5 and/or
DQ2.2) and -DQ8 using tests of EUROIMMUN AG (Lübeck, Germany). In sera only
positive for IgG-aDGL, total IgA was nephelometrically measured (Roche, Mannheim,
Germany). IgA values of ≤ 0.05 g/l were considered as sign of selective IgA
deficiency (sIgAD).
In a third step, available follow-up sera (T1) of patients with conspicuous results for
IgA-aTTG and/or IgG-aDGL (HLA-DQ2 and/or DQ8 positive or HLA status not
known) were analysed for IgA-aTTG, IgG-aDGL and EmA. Finally, we invited
participants (T2) who showed positivity for either IgA-aTTG and/or IgG-aDGL at T0
and T1. Cases with negative HLA results were not pursued.
The final step included a personal interview which comprises questions concerning
gastrointestinal complaints and further CD-relevant symptoms, associated diseases,
family history and gluten uptake as well as the decision of the participant for further
clarification. The interview was performed by an experienced paediatric
gastroenterologist. The latter is still ongoing.
Interim analysis
To compare our data with other screening studies, the following groups were
considered:
Known CD (group 1): known CD with HLA-DQ2/DQ8 positivity.
Seropositivity (group 2): IgA-aTTG >1 x ULN or (IgG-aDGL >1 x ULN and
sIgAD) + group 1.
Potential CD (group 3): (IgA-aTTG >1 x ULN and IgA-EmA-positive) or (IgGaDGL >1 x ULN and sIgAD and HLA-positive) + group 1.
Probable CD (group 4): IgA-aTTG ≥10 x ULN and IgA-EmA-positive + group 1.
Results and discussion
Characteristics of our screening cohort are shown in Table 1. Results regarding the
first two steps of our screening procedure are depicted in Fig. 2. Of 2363 children and
adolescents, 29 were only positive for IgA-aTTG (1.23%) and 23 only for IgG-aDGL
(0.87%). The results for IgA-aTTG are comparable with a previous observation in an
adult cohort [3]. Double positivity for the indicated antibodies was observed for 11
patients (0.47%, not shown). Of seven participants who noted that CD was previously
diagnosed, one was positive for IgA-aTTG (probable under GFD) but only four
(including the indicated patient) were HLA-DQ2 and/or DQ8 positive. We conclude
that inclusion of probands indicating to have CD into the calculation of prevalence
without HLA-typing [4] leads to an overestimation of frequency.
Table 1. Baseline demographics and clinical characteristics.
Figure 2. Screening results regarding IgA-aTTG, IgG-aDGL, IgA-EmA and HLA-type.
HLA+ HLA-DQ2 and/or DQ8 positive, IgA-EmA+ IgA-EmA positive, sIgAD selective
IgA deficiency, ULN upper limit of normal
None of the participants with solely IgG-aDGL positivity was positive for IgA-EmA.
Otherwise, all participants with IgA-aTTG ≥5 x ULN were also positive for the
immunofluorescence test. In the group of probands with IgA-aTTG between 1 and 5 x
ULN, there were only eight of 13 sera in which IgA-EmA were detected. Further, none
of the sixteen patients (0.67%) with IgA-aTTG between 0.5 and 1 x ULN show IgAEmA
positivity. These findings contradict the observations of a large Swedish
screening study in which 25% of the IgA-TTG negative probands (between 0.5 and 1 x ULN) were IgA-EmA positive [8]. This is possibly due to the use of a lower cut-off
for IgA-EmA (1:5) or, more likely, to a low sensitivity of the ELISA test.
Finally, positive antibody tests were verified by HLA-genotyping if DNA was
available. All participants with IgA-EmA positivity were either HLA-DQ2 and/or
DQ8 suggesting that HLA-typing is not necessary if patients show IgA-EmA
positivity. Otherwise, probands with IgA-aTTG between 1 and 5 x ULN without IgAEmA
positivity were only positive for HLA-DQ2/8 in four of seven cases.
Furthermore, patients positive for only IgG-aDGL have a false-positive rate of 50%
indicating the necessity for HLA-typing in these cases. This supports the
recommendation to genotype IgA-aTTG seronegative patients with CD-related signs
[9] to evaluate the need of subsequent endoscopic evaluation. In case of HLA-DQ2/8
negativity CD seems to be very unlikely and other differential diagnoses have to be
considered. We found one participant with sIgAD who was positive for IgG-aDGP
carrying HLA-DQ2.
Summarising, we obtained the following frequencies:
Group 1 (known CD) = 0.16% (CI95% 0.04-0.41)
Group 2 (seropositivity) = 1.39% (CI95% 0.97-1.98)
Group 3 (potential CD) = 1.06% (CI95% 0.70-1.58)
Group 4 (probable CD) = 0.51% (CI95% 0.28-0.91)
In comparison to the most recent German prevalence study [4], the proportion of
children being seropositive or having known CD (sum of groups 1 and 2) is higher in
our screening study (1.39% vs. 0.9%). As expected, the number of probands in group 3
is lower than in group 2. This is mainly due to sera with IgA-aTTG <5 x ULN but
IgA-EmA negative. Potential CD is actually defined as IgA-aTTG, IgA-EmA and
HLA-DQ2/8 positivity without severe intestinal impairment (≤ Marsh 1). Due to the
fact that all EmA-positives were also HLA-DQ2/DQ8 positive we put back HLAtyping
for discussion. Omitting HLA-typing in case of IgA-EmA positivity in addition
to IgA-aTTG >1 x ULN seems meaningful for the clinical application to reduce costs.
This contradicts current diagnostic recommendations [10] for symptomatic patients
with IgA-aTTG ≥10 x ULN suggesting confirmatory assays of IgA-EmA and HLA.
Children in group 4 seem have CD with high probability [6]. It was shown that IgAaTTG
levels ≥10 x ULN predict mucosal damage ≥ Marsh 3A in a population basedscreening
independently from the patient’s symptomatic state. Contrary to our
findings, the proportion of probands with IgA-aTTG ≥10 x ULN was higher (0.48%
vs. 0.33%). In addition, Laass et al. [4] found a frequency of 0.4% for this group of
seropositive probands but concluded that all seropositives and those with known CD
(without HLA confirmation) reflect the actual prevalence in Germany. Interestingly,
the frequency of group 4 exceeds the estimated German prevalences for biopsy-proven
CD [1,2].
We obtained 41 of 61 requested follow-up sera including those from children with
associated diseases. We observed seroconversion from positive to negative for six
participants (Fig. 3A). Interestingly, these sera had an initial IgA-aTTG value between
0.5 and 1 x ULN. One was also positive for IgG-aDGL. All except one were HLADQ2/
DQ8 positive. Of these, only one was IgA-EmA positive. Concerning these
results, there is evidence that there is a fraction of probands with suspected CD that
should be followed-up if initial IgA-aTTG values are between 0.5 and 1xULN. In
contrast, Fig. 3B represents IgA-aTTG values of participants which decreased from
positive values to concentrations below the threshold at follow-up. One was a patient
with known CD and the decrease can be attributed to a GFD. For the other two, we
have currently no information on possible reasons (GFD or false positive at T0).
Figure 3. Seroconversions of IgA-aTTG during follow-up (T1). A) Seroconversion
from negative to positive B) Seroconversion from positive to negative
Conclusion
The prevalence of CD in the LIFE Child study cohort ranges from 0.16 to 1.39%
depending on the kind of calculation/estimations and exceeds previous observations.
The percentage of patients with highly probable CD is 0.51%. Calculation of CD
prevalence relying only on antibody values above manufacturer’s cut-off without
further confirmation by EmA or HLA testing results in overestimation. Cases of socalled “already known” CD have to be considered with caution and should be
controlled by genotyping. Finally, children with IgA-aTTG values between 0.5 and 1 x
ULN should be followed up. We are currently trying to clarify the cases of suspected
CD in our screening. For that purpose, the participants are invited for an individual
interview and, if necessary, we recommend an appointment with a paediatric
gastroenterologist.
Acknowledgements
The study was partially funded by the research fund of the Deutsche
Zöliakiegesellschaft. We also thank EUROIMMUN AG (Conny Daehnrich, Annika
Jahnke, Kathrin Axel und Ulf Steller) for providing the ELISA kits and analysing
EmA and HLA-DQ2/Q8.
References
1. Kratzer W. Prevalence of celiac disease in Germany: A prospective follow-up
study. WJG 2013; 19: 2612-2620.
2. Henker J. Prävalenz der asymptomatischen Zöliakie bei Kindern und
Erwachsenen. Dtsch Med Wochenschr 2002; 127: 1511-1515.
3. Metzger MH, Heier M, Maki M, et al. Mortality excess in individuals with
elevated IgA anti-transglutaminase antibodies: the KORA/MONICA Augsburg
cohort study 1989-1998. Eur J Epidemiol 2006; 21: 359-365.
4. Laass MW, Schmitz R, Uhlig HH, et al. The prevalence of celiac disease in
children and adolescents in Germany. Deutsches Arzteblatt international 2015;
112: 553-560.
5. Mustalahti K, Catassi C, Reunanen A, et al. The prevalence of celiac disease in
Europe: results of a centralized, international mass screening project. Ann Med
2010; 42: 587-595.
6. Webb C, Norstrom F, Myleus A, et al. Celiac disease can be predicted by high
levels of anti-tissue transglutaminase antibodies in population-based screening. J
Pediatr Gastroenterol Nutr 2015; 60: 787-791.
7. Quante M, Hesse M, Dohnert M, et al. The LIFE child study: a life course
approach to disease and health. BMC Public Health 2012; 12: 1021.
8. Sandstrom O, Rosen A, Lagerqvist C, et al. Transglutaminase IgA antibodies in a
celiac disease mass screening and the role of HLA-DQ genotyping and endomysial
antibodies in sequential testing. J Pediatr Gastroenterol Nutr 2013; 57: 472-476.
9. Volta U, Caio G, Boschetti E, et al. Seronegative celiac disease: Shedding light on
an obscure clinical entity. Dig Liver Dis 2016; 48: 1018-1022.
10. Husby S, Koletzko S, Korponay-Szabó I, et al. European Society for Pediatric
Gastroenterology, Hepatology, and Nutrition Guidelines for the diagnosis of
coeliac disease. J Pediatr Gastroenterol Nutr 2012; 54: 136-60.
Results of the prospective multicentre trial of
antibody diagnostics in coeliac disease (AbCD)
Johannes Wolf1, David Petroff2, Dirk Hasenclever3, Thomas Mothes1, and the AbCDstudy
group: Thomas Richter4, Marcus Auth5, Holm Uhlig6, Martin Laass7, Klaus-
Michael Keller8, Andreas Krahl9, Norman Händel10, Klaus-Peter Zimmer11, Almuthe
Hauer12, Matthias Heiduk13, Gunter Flemming14, Frank Schmidt15
1 Institute of Laboratory Medicine, Clinical Chemistry and Molecular Diagnostics,
Medical Faculty of the University and University Hospital, Leipzig, Germany
2 Coordination Centre for Clinical Trials of the University, Leipzig, Germany
3 Institute of Medical Informatics, Statistics and Epidemiology of the University,
Leipzig, Germany
4 Children‘s Hospital of the Clinical Centre ‘‘Sankt Georg’’, Leipzig, Germany
5 Alder Hey Children’s National Health Service Foundation, Liverpool, UK
6 Translational Gastroenterology Unit, Experimental Medicine, University of Oxford,
John Radcliffe Hospital, Oxford, UK
7 University Children‘s Hospital, Dresden, Germany
8 Children‘s Hospital, DKD Helios Medical Centre, Wiesbaden, Germany
9 Children‘s Hospital „Prinzessin Margaret“, Darmstadt, Germany
10 University Children‘s Hospital, Leipzig, Germany
11 University Children‘s Hospital, Gießen, Germany
12 University Children‘s Hospital, Graz, Austria
13 Department of Paediatrics, Helios Hospital, Plauen, Germany
14 Children‘s Hospital of the Medical School, Hannover, Germany
15 University Children‘s Hospital, Halle, Germany
Introduction
Diagnosis of coeliac disease (CD) relies on a combination of clinical, genetic,
serological and duodenal morphological findings. The European Society for Paediatric
Gastroenterology, Hepatology, and Nutrition (ESPGHAN) published new guidelines
in the year 2012 that permitted diagnosis of CD without biopsies in symptomatic
patients with a concentration of IgA antibodies to tissue transglutaminase (aTTG) ≥10
times the upper limit of normal (10×ULN) upon confirmation with assay of IgAendomysium
antibodies (EMA) and HLA-typing, but noted that this needs to be
evaluated prospectively [1].
A retrospective study proposed two diagnostic procedures based on (a) IgA-aTTG and
total IgA (TTG/IgA-procedure) and (b) IgA-aTTG and IgG antibodies to deamidated
gliadins (TTG/DGL-procedure). This study uses both one- and tenfold ULN, resulting
in the classification “CD”, “non-CD” or “biopsy required” [2].
A search in PubMed revealed that until now there was only one paediatric prospective
study with 183 children in which the “10×ULN rule” did not lead to any false-positive
findings [3]. The usefulness of another antibody species, IgG-antibodies against
deamidated gliadin peptides (aDGL) is still under discussion.
The prospective multicentre AbCD study was performed to validate positive / negative
predictive values (PPV / NPV) of these procedures.
Materials and methods
Thirteen centres from Germany, the United Kingdom and Austria enrolled paediatric
patients scheduled for duodenal biopsy to confirm or rule out CD. Antibodies were
measured blinded by a laboratory (EUROIMMUN, Dassow, Germany) and tissue
sections underwent reference pathology. After follow-up, the paediatricians made a
diagnosis largely following routine clinical procedures.
We defined two antibody procedures as follows:
TTG/IgA-procedure (sIgAD excluded):
CD if IgA-aTTG ≥10×ULN;
no CD if IgA-aTTG <1×ULN;
otherwise: biopsy necessary.
TTG/DGL-procedure:
CD if IgA-aTTG ≥10xULN or IgG-aDGL ≥10×ULN;
no CD if IgA-aTTG <1xULN and IgG-aDGL <1×ULN;
otherwise: biopsy necessary.
The trial was registered on the Internet Portal of the German Clinical Trials Register
(https://drks-neu.uniklinik-freiburg.de/drks_web/setLocale_EN.do; DRKS00003854).
Results and discussion
For 898 of 949 participants, serum, biopsy, and follow-up data were available (592
CD, 345 non-CD, 24 no final diagnosis). A strength of our trial is, that we did not
exclude the 24 patients in whom the diagnostic process did not lead to a final
diagnosis, who are commonly neglected in diagnostic studies. These patients were
included into our calculation of PPV and NPV as false-positive or false-negative cases.
The TTG/DGL-procedure, based on the assay of IgA-aTTG and IgG-aDGL can safely
diagnose (IgA-aTTG or IgG-aDGL ≥10×ULN) or exclude CD (IgA-aTTG and IgGaDGL <1×ULN) irrespective of the presence of symptoms in more than three quarters
of children and adolescents (positive as well as negative predictive values >95%). The
TTG/IgA-procedure, based on the assay of IgA-aTTG and of total IgA (IgA-aTTG ≥10×ULN or <1×ULN for IgA-competent patients) is almost identical in its ability to
diagnose CD, but has a slightly narrower prevalence range over which it can safely exclude CD. The lower limit of 95% confidence bound (LCB) is above 0.9 for each
procedure.
Calculation of predictive values for the subset of our patients with a final diagnosis
results in even higher predictive values.
Model based extrapolation shows that PPV and NPV remain above 0.95 with LCB> 0.9 even at a prevalence as low as 4%. Endomysium antibodies and HLA-typing did
not improve PPV in samples with IgA-aTTG ≥10×ULN. Notably, the discrepancy rate
between pathologists is 4.2% and comparable to the error rate in the serological
procedures.
Conclusion
AbCD is the first prospective multi-centre trial providing evidence on the reliability of
antibody tests including IgG-aDGL in diagnostics of CD based on a large number of
children and adolescents. Our results have major personal and health care implications
in clinical practice. They demonstrate that duodenal biopsy can be avoided in three
quarters of paediatric patients with suspected CD, thereby reducing costs, endoscopy
waiting times, patient risks, and delay of treatment. Furthermore, our study shows that
other diagnostic tests such as HLA-typing or assay of EMA are not required in
unequivocal cases.
References
1. Husby S, Koletzko S, Korponay-Szabó I, et al. European Society for Pediatric
Gastroenterology, Hepatology, and Nutrition Guidelines for the diagnosis of
coeliac disease. J Pediatr Gastroenterol Nutr 2012; 54: 136-60.
2. Wolf J, Hasenclever D, Petroff D, et al. Antibodies in the diagnosis of coeliac
disease: A biopsy-controlled, international, multicentre study of 376 children with
coeliac disease and 695 controls. PLoS ONE 2014; 9: e97853.
3. Mubarak A, Wolters VM, Gmelig-Meyling FHJ, et al. Tissue transglutaminase
levels above 100 U/mL and celiac disease: a prospective study. World J
Gastroenterol 2012; 18: 4399-403.
6 Enzymatic gluten degradation
Identification of novel and food-grade glutendegrading
enzymes
Eva J. Helmerhorst1, Guoxian Wei1, Na Tian1, Detlef Schuppan2
1 Department of Molecular and Cell Biology, Boston University Henry M. Goldman
School of Dental Medicine, Boston, MA, USA
2 Institute of Translational Immunology and Research Center for Immunology,
University Medical, Center, Johannes-Gutenberg-University, Mainz, Germany
Introduction
Gluten proteins have received much attention ever since it was discovered that they are
the causative agents in coeliac disease (CD), a T-cell mediated inflammatory disorder
of the small intestine [1-4]. Despite the discovery of the gluten-CD causal relationship
in the 1950s [5], there is still no pharmacological therapy for CD, and individuals
diagnosed must follow a strict life-long gluten-free diet. Several factors contribute to
the toxicity of gluten proteins for CD patients. One critical feature of gluten proteins is
that the domains comprising the immunogenic epitopes resist degradation by
mammalian digestive enzymes, and thus are not neutralised during gastro-duodenal
transport [6]. Luminal enzyme therapeutic approaches aim to abolish gluten with
exogenous enzymes exhibiting cleavage specificities in immunogenic epitopes [7-11].
The immunogenic gluten epitopes all contain glutamine residues that can be
selectively deamidated by tissue transglutaminase, and have consecutive proline (P)
and glutamine (Q) residues in their sequences [12]. Our research on the peptidome of
human saliva “accidentally” identified an enzyme that hydrolyses the XPQ sequence
after Q in salivary proline-rich proteins [13]. This paper summarises the isolation of
the gluten-degrading oral species from mixed culture biofilm, and the isolation of the
enzyme. The discovery of a food-grade natural variant of this enzyme [14] offers novel
and readily available therapeutic perspectives for patients suffering from gluten
intolerance disorders, including CD.
Materials and methods
Dental plaque was collected according to a protocol approved by the Institutional
Review Board for human subjects research at Boston University. The plaque was
suspended in phosphate-buffered saline and plated on blood agar containing haemin
and vitamin K. Individual colonies were then subcultured on agar plates containing
wheat gluten as the sole protein source. Strains growing well on this agar were further
subcultured, alternatingly on blood agar and gluten agar, to purity. Microbe-associated
enzyme activities were evaluated in four enzyme assays. In the first, a series of paranitroanilide-derivatised gluten-derived tripeptide enzyme substrates were
employed. In the second assay, gliadin-degrading activity was investigated in-gel in a
gliadin zymogram assay. Third, anti-gliadin activity in solution was studied by SDSPAGE.
Lastly, the enzymatic activity towards the immunogenic, protease-resistant 33-
mer peptide was investigated by RP-HPLC. Strains of interest were identified by 16S
rDNA analysis. The gluten-degrading enzyme from R. mucilaginosa was isolated from
cells treated with lysozyme and a mild detergent, followed by cell sonication,
ultracentrifugation, DEAE-chromatography, and gliadin zymography. Enzyme-active
bands were excised, trypsinised, and subjected to LC-ESI-MS/MS for protein
identification. Epitope abolishment by the purified Rothia enzyme as well as
commercially available Bacillus enzymes was investigated using ELISA assays
employing the R5 and G12 antibodies. The experimental details of the above
experiments have been published [14-19].
Results and discussion
Mixed gliadins, incubated in a suspension of dental plaque bacteria, were degraded,
and the half-life of 250 μg/ml gliadins in a suspension with an OD620 of 1.2 was
determined to be 6 h [17]. The isolation of the gluten-bacterial species among the over
1,000 different microbial species contained in dental plaque [20] was accomplished by
a selective gluten agar approach [16,19]. Several strains grew well and were then
evaluated in the enzyme assays employing gluten-derived substrates. The strains active
in at least two of the four assays were speciated as Rothia aeria, Rothia mucilaginosa,
Actinomyces odontolyticus, Neisseria mucosa, Capnocytophaga sputigena, and two
Streptococcus species [15,19]. Among these, the Rothia bacteria were active in all four
enzyme assays. Suspensions of R. aeria, or R. mucilaginosa, degraded mixed gliadins
very rapidly, at a 25-fold increased rate compared to mixed dental plaque suspension.
The tripeptide substrates hydrolysed most rapidly were Z-YPQ-pNA and Z-LPY-pNA.
The apparent molecular weights of the enzymes from R. mucilaginosa and R. aeria
were between 70,000 - 80,000. The optimum pH for activity was in the neutral pH
range, with R. aeria showing residual activity at pH values as low as 3.0 [19]. We
confirmed earlier reports [6] that the immunogenic 33-mer gliadin peptide was entirely
resistant to pepsin, trypsin and chymotrypsin (Fig. 1). The 33-mer, however, was
completely degraded by Rothia-associated enzymes, within 2 h of incubation (Fig. 1).
The sequenced degradation fragments showed evidence for preferential cleavage
activity after XPQ↓ and XPY↓, consistent with the results obtained with the synthetic
tripeptide substrates [18,19]. The enzyme was subsequently isolated from R.
mucilaginosa, for which the full genomic sequence was available at the time. The
enzyme was found to be cell-bound, and therefore was extracted from a cell sonicate
[14]. The proteins in the extract were separated by DEAE anion-exchange
chromatography. Proteins with enzymatic activity eluted early in the chromatogram
and were thus mildly anionic in nature. The proteins contained in these fractions
exhibited molecular weights of 125,000, 75,000 and 80,000. Enzyme activity was associated with the 75,000 and 80,000 bands, but not with the 125,000 band, as
revealed by zymography. The excised and in-gel trypsinised bands (75,000, 80,000
and 125,000) were analysed by mass spectrometry. This led to the identification of a
mixture of the hypothetical proteins C6R5V9 and C6R5W1 in the inactive 125,000
band, C6R5V9 in the active 80,000 band, and C6R5W1 in the active 75,000 band.
Figure 1. Degradation of the 33-mer gliadin peptide by mammalian enzymes (left
panels) or oral microbial enzymes associated with oral strain WSA-8 (right panel).
The 33-mer peptide was dissolved to 250 μg/mL and incubated with 1 μg/mL
chymotrypsin, pepsin or trypsin (left panels) or in a suspension of oral microbial
strain WSA-8 (R. aeria; OD620=1.2). The incubation buffer composition mimicked the
ion composition of human saliva (pH 7.0). Sample aliquots were removed at t=0 and
t=24 h (left panels) or t=0, 3 min, 60 min and 120 min (right panel). Degradation of
the 33-mer in incubation aliquots was monitored by RP-HPLC
From this result it was concluded that the full length C6R5V9 and C6R5W1 proteins
in the 125,000 band are proteolytically processed to yield the active 80,000 and 75,000
enzymes, respectively. Structural analysis of the primary sequences revealed that both
proteins are members of the subtilisin enzyme family [14]. The Rothia subtilisins are
about 3-fold larger in size than the well-known and structurally resolved Bacillus
subtilis-derived subtilisins, and show very little similarity in primary amino acid
sequences. Nevertheless, like Rothia, the Bacillus subtilisins, specifically subtilisin A,
and nattokinase, cleaved gliadins rapidly [14]. Rothia subtilisins eliminated both the R5 and G12 epitopes, while the Bacillus subtilisins primarily abolished the R5 epitope
(Fig. 2). This indicates some differences in substrate preference among the Bacillus
and Rothia subtilisins that could be clinically significant. The theoretical cleavage
specificity of subtilisins, XPX↓, matches the experimentally observed cleavage of the
33-mer at the XPQ↓ and XPY↓ positions. Importantly, these domains are contained in
all known gliadin-derived immunogenic epitopes.

Figure 2. Gliadin epitope abolishment by subtilisins. Mixed gliadins were incubated
with R. mucilaginosa enzyme preparation (Rmep) enriched in subtilisins. Aliquots
were analysed for the survival of epitopes QQPFP in the R5 ELISA assay (A) and
QPQLPY in the G12 ELISA assay (B). Results obtained with B. subtilis subtilisin A as
the enzyme source are shown in C and D. Data adapted from Wei et al., 2016 [14]
Conclusion
The subtilisins isolated from non-pathogenic oral Rothia bacteria showed a remarkable
activity towards mixed gliadins and capacity to neutralise the most immunogenic
epitopes implicated in CD. What makes these enzymes of particular interest is the fact
that certain members of the subtilisin family are already food-grade, e.g. nattokinase, which demonstrated significant gluten-degrading activities as well. The latter enzyme
is present in the Bacillus-containing food product called natto which is widely
consumed in Japan. Coincidentally, the prevalence of CD in these regions is low. Our
discoveries have opened new therapeutic avenues for the further development of the
subtilisin enzymes as dietary supplements to help in the digestion of gluten in vivo.
References
1. Abadie V, Sollid LM, Barreiro LB, et al. Integration of genetic and immunological
insights into a model of celiac disease pathogenesis. Annu Rev Immunol 2011; 29:
493-525.
2. Green PH, Jabri B. Celiac disease. Annu Rev Med 2006; 57: 207-221.
3. Schuppan D, Junker Y, Barisani D. Celiac disease: from pathogenesis to novel
therapies. Gastroenterology 2009; 137: 1912-1933.
4. Koning F, Schuppan D, Cerf-Bensussan N, et al. (2005). Pathomechanisms in
celiac disease. Best Pract Res Clin Gastroenterol 2005; 19: 373-387.
5. van Berge-Henegouwen GP, Mulder CJ. Pioneer in the gluten free diet: Willem-
Karel Dicke 1905-1962, over 50 years of gluten free diet. Gut 1993; 34: 1473-
1475.
6. Shan L, Molberg O, Parrot I, et al. (2002). Structural basis for gluten intolerance
in celiac sprue. Science 2002; 297: 2275-2279.
7. Gass J, Bethune MT, Siegel M, et al. Combination enzyme therapy for gastric
digestion of dietary gluten in patients with celiac sprue. Gastroenterology 2007;
133: 472-480.
8. Lähdeaho ML, Kaukinen K, Laurila K, et al. Glutenase ALV003 attenuates gluteninduced
mucosal injury in patients with celiac disease. Gastroenterology 2014;
146: 1649-1658.
9. Stepniak D, Spaenij-Dekking L, Mitea C, et al. Highly efficient gluten degradation
with a newly identified prolyl endoprotease: implications for celiac disease. Am J
Physiol Gastrointest Liver Physiol 2006; 291: G621-G629.
10. Tack GJ, van de Water JMW, Bruins MJ, et al. Consumption of gluten with
gluten-degrading enzyme by celiac patients: a pilot-study. World J Gastroenterol
2013; 19: 5837-5847.
11. Mitea C, Havenaar R, Drijfhout JW, et al. Efficient degradation of gluten by a
prolyl endoprotease in a gastrointestinal model: implications for coeliac disease.
Gut 2008; 57: 25-32.
12. Sollid LM, Qiao SW, Anderson RP, et al. Nomenclature and listing of celiac
disease relevant gluten T-cell epitopes restricted by HLA-DQ molecules.
Immunogenetics 2012; 64: 455-460.
13. Helmerhorst EJ, Sun X, Salih E, et al. Identification of Lys-Pro-Gln as a novel
cleavage site specificity of saliva-associated proteases. J Biol Chem 2008; 283:
19957-19966.
14. Wei G, Tian N, Siezen R, et al. Identification of food-grade subtilisins as glutendegrading
enzymes to treat celiac disease. Am J Physiol Gastrointest Liver Physiol
2016; 311: G571-G580.
15. Fernandez-Feo M, Wei G, Blumenkranz G, et al. The cultivable human oral
gluten-degrading microbiome and its potential implications in coeliac disease and
gluten sensitivity. Clin Microbiol Infect 2013; 19: E386-E394.
16. Helmerhorst EJ, Wei G. Experimental strategy to discover microbes with glutendegrading
enzyme activities. Proc. SPIE 2014; 9112, 91120D1-11.
17. Helmerhorst EJ, Zamakhchari M, Schuppan D, et al. (2010). Discovery of a novel
and rich source of gluten-degrading microbial enzymes in the oral cavity. PLoS
One 2010; 5: e13264.
18. Tian N, Wei G, Schuppan D; et al. Effect of Rothia mucilaginosa enzymes on
gliadin (gluten) structure, deamidation, and immunogenic epitopes relevant to
celiac disease. Am J Physiol Gastrointest Liver Physiol 2014; 307: G769-G776.
19. Zamakhchari M, Wei G, Dewhirst F, et al. (2011). Identification of Rothia bacteria
as gluten-degrading natural colonizers of the upper gastro-intestinal tract. PLoS
One 2011; 6: e24455.
20. Dewhirst FE, Chen T, Izard J, et al. (2010). The human oral microbiome. J
Bacteriol 2010; 192: 5002-5017.
Studying kinetics of intestinal gluten degradation
using peptide libraries
Niels Röckendorf, Andreas Frey
Division of Mucosal Immunology & Diagnostics, Priority Area Asthma and Allergy,
Research Center Borstel, Borstel, Germany
Introduction
In Western countries about 1% of the population suffer from coeliac disease (CD) [1-
3], a multifactorial chronic inflammatory intestinal disorder that is triggered by cereal
storage proteins, the so-called gluten. Gluten consists of gluey, poorly water-soluble,
proline-rich polypeptides that constitute about 10% of the mass of cereal flour and are
important for the baking process and the taste, texture and appearance of the resulting
product [4,5]. Although consumption of gluten is a necessary requirement for the
development, manifestation and progression of CD, it alone is not sufficient to cause
illness. Additional factors must coincide to boost disease [6]. On the genetic side, this
applies mainly to the presence of major histocompatibility genotypes HLA DQ2 or
DQ8, as almost all affected individuals express one of these haplotypes [7]. It was
shown that peptidic fragments of the gluten fraction avidly bind in particular to these
MHC-class II variants giving rise to proinflammatory T cells which in turn foster
disease and its persistence [7]. Yet, with a prevalence of about 1% for CD and about
30-40% of HLA DQ2/8 present in the population [1-3] additional environmental or
physiological elicitors must exist. Such a factor may be the gastrointestinal degradome
and its capacity to cleave gluten peptides, in particular the ones that bind to HLA
DQ2/8. If those peptides can be broken down completely before they reach their target
antigen-presenting cells in the alimentary tract, gluten-specific T cells cannot be
generated and disease cannot develop, even if an individual is HLA DQ2/8 positive.
As a comprehensive degradation profile of CD-active gluten peptides in oro-gastrointestinal
fluids does not exist we wanted to address this question using a recently
developed peptide stability assay [8]. A major advantage of the assay in this context is
its robustness regarding the proteases and peptides to be used and the respective
readout it creates. The assay accepts crude protease mixtures such as gastrointestinal fluids, accommodates every conceivable amino acid sequence motif and yields true
half-lifes - as long as the stability of the substrate peptide is not orders of magnitude
higher than that of the protease mixture itself. In our first round of experiments
reported herein we used rat small intestinal juice as model protease mixture [9] and a
battery of T-cell epitope-sized gluten peptides whose amino acid sequence was shown
to “elicit coeliac disease or to activate MHC class II restricted T cells of subjects with
coeliac disease” [10,11] as substrates. The data shall provide information on the
feasibility of such a study on human beings and provide a first clue about the gluten peptide and T-cell epitope half-lives to be expected after exposure to crude intestinal
juices.
Materials and methods
A compilation of CD-active gluten sequence motifs was procured from the
AllergenOnline database [11]. The selected sequences were synthesised by solid phase
peptide synthesis, extracted from the solid support and subjected to proteolysis testing
as described in detail by Gorris et al. [8]. Briefly, proline-functionalised cellulose
sheets were equipped in a spatially resolved manner with Boc-Lys (Fmoc) to provide
an acid-cleavable anchor onto which a Lys-ɛ-biotin - polyethylene glycol - Ddiglutamate “detection-insulator-unit” was positioned. Thereafter, the respective
peptides were synthesised onto these areas using traditional Fmoc solid phase peptide
synthesis before, in a final step, another “insulator-capture-unit” composed of Ddiglutamate
- polyethylene glycol - aminoundecanoic acid - 2,4-dichlorophenoxyacetic
acid was attached on each spot. The synthesis areas were punched out and the residing
double-tagged peptides were deprotected and cleaved-off by acid treatment. The crude
peptides can be used without further purification in the subsequent digestion assay.
Intestinal juice was harvested from female Wistar rats of 6 weeks of age in undiluted
form using absorbent filter wicks [9], eluted by centrifugation and pooled. Animal
experiments were approved by the Ministry of Energy, Agriculture, the Environment
and Rural Areas of Schleswig-Holstein, Kiel, FRG [No. V312-7224.123-3]. The tota
proteolytic activity of the intestinal fluid was determined using a commercially
available azocasein proteolysis assay (Sigma #A2765, Sigma-Aldrich, Steinheim,
FRG; ref. [12]). For proteolysis testing of gluten peptides, the intestinal fluid was
serially diluted with buffer mimicking the conditions of the small intestine in terms of
pH and ionic strength in microplates. Peptide solutions were added and the mixtures
were allowed to react for defined periods of time at 37 °C. The reactions were
terminated by addition of protease inhibitors and heating and the respective amounts of
remaining intact peptide were determined in a microplate assay using anti-2,4-
dichlorophenoxyacetic acid antibodies as capture and streptavidin-peroxidase as
detection reagents. Due to the minute amounts of peptide-substrate used (≤ 0.75 pMol
per digestion reaction) the kinetics follow pseudo-first-order and the results can be
reported as peptide half-lives in undiluted intestinal juices. Data analysis was
performed with the Prism v5.0f software package, GraphPad, Ja Jolla, CA, USA.
Results and discussion
A total of 458 CD-active 8-15mer peptides derived from wheat, rye, barley, and oat
proteins were tested for their cleavability by rat small intestinal fluid under
physiological conditions. As readouts half-lives in rat small intestinal juice at 37 °C
were recorded for each peptide. Since reporting all individual readouts tabularly would
be of little value to convey our findings we refrain from listing those data in this report. They will be made available to interested researchers from the authors upon
request1.
Under our experimental conditions, the half-lives of the CD-active cereal protein
sequence motifs range from 2 ms to 20 min with a mean value of approximately 8 s
(Fig. 1). Although these values seem to indicate rapid digestion, this is actually not the
case when juxtaposed with fragments of other typical dietary proteins. Comparison of
our data with the half-lives of chicken egg ovalbumin peptides in murine small
intestinal juice, which we have determined in a previous work [8], highlights this
point. While we could establish that the total proteolytic activities of the mouse and
the rat small intestinal juices were comparable, the half-lives of 16mer ovalbuminderived
peptides in murine small intestinal juice were considerably lower than those of
the gluten peptides in rat intestinal juice, ranging from 800 μs to 6.4 s with a mean
value of 152 ms. Even shorter, less proteolysis-prone 10mer ovalbumin peptides
display a mean half-life of only 1.3 s (range 800 μs to 40 s) (Fig. 1). In a second round
of analysis we focused on CD-relevant gluten epitopes of which the specific T-cell
reactivity has been defined in detail, namely those CD-active peptides which contain
one of the 9mer core, DQ2- or DQ8-resticted T-cell epitopes compiled by Sollid and
colleagues [13]. A total of 112 peptides fulfilled this criterion. Here the mean half-life
was slightly lower (5 s) than that of the total pool tested but the overall distribution
profile was very similar (Fig. 1).
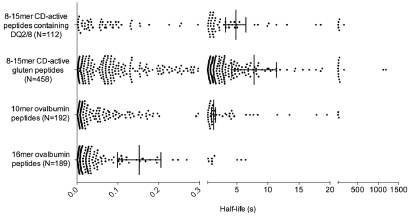
Figure 1. Comparison of the stability of CD-active gluten peptides and peptides
derived from chicken egg ovalbumin in rodent small intestinal juice. Individual halflives
of all peptides analysed are depicted. Vertical bars indicate mean ± SEM
Thus, the CD-active gluten peptides analysed are significantly (p < 0.05, Kruskal-
Wallis test with Dunn’s Multiple Comparison post hoc test) more stable than those of
ovalbumin when tested in the rodent system. Still, when one considers the typical
gastrointestinal transit times for rodents of about 90 min to 2 h [14-16] only the most
stable gluten peptides should be able to pass the intestines to a considerable extent
without being broken down, and therefore may access every cell type of the mucosal
lining. The majority of potential MHC class II-restricted gluten T-cell epitopes will not
survive transit. This might be an explanation for the fact that rodents do not develop
CD-like enteropathy, even if expressing HLA-DQ2 and gluten-specific T-cell
receptors [17], although additional factors promoting disease in the human system may
also be missing in those models.
Conclusion
The results presented above confirm earlier findings on the high stability of cereal
storage proteins and show that even the regular small intestinal degradome of rodents
is not ideally suited to cope with cereal storage proteins and to completely clear
gluten-borne T-cell epitopes. It will be of great interest to port this assay to the human
system and investigate whether in humans, differences in the proteolytic stability of
CD-active peptides in the intestine might correlate with a disposition for CD.
References
1. Lionetti E, Gatti S, Pulvirenti, et al. Celiac disease from a global perspective. Best
Pract Res Clin Gastroenterol 2015; 29: 365-379.
2. Catassi C, Gatti S, Lionetti E. World perspective and celiac disease epidemiology
Digest Dis 2015; 33: 141-146.
3. Lionetti E, Catassi C. Co-localization of gluten consumption and HLA-DG2 and– DQ8 genotypes, a clue to the history of celiac disease. Digest Liver Dis 2014; 46:
1057-1063.
4. Scherf KA, Koehler P, Wieser H. Gluten and wheat sensitivities – an overview. J
Cereal Sci 2016; 67: 2-11.
5. Delcour JA, Joye IJ, Pareyt B, et al. Wheat gluten functionality as a quality
determinant in cereal-based food products. Annu Rev Food Sci Technol 2012; 3:
469-492.
6. Green PHR, Lebwohl B, Greywoode R. Celiac disease. J Allergy Clin Immunol
2015; 135: 1099-1106.
7. Qiao S-W, Sollid LM, Blumberg RS. Antigen presentation in celiac disease. Curr
Opin Immunol 2009; 21: 111-117.
8. Gorris HH, Bade S, Röckendorf N, et al. Rapid profiling of peptide stability in
proteolytic environments. Anal Chem 2009; 81: 1580-1586.
9. Bade S, Gorris HH, Koelling S, et al. Quantitation of major protein constituents of
murine intestinal fluid. Anal Biochem 2010; 406: 157-165.
10. Goodman RE, Ebisawa M, Ferreira F, et al. AllergenOnline: A peer-reviewed,
curated allergen database to assess novel food proteins for potential crossreactivity.
Mol Nutr Food Res 2016; 60: 1183-1198.
11. Food Allergy Research and Resource Program (FARRP), AllergenOnline database
(v14.0), Celiac Disease Peptide and Protein Database. www.allergenonline.org
12. Charney J, Tomarelly RM. A colorimetric method for the determination of the
proteolytic activity of duodenal juice. J Biol Chem 1947; 171: 501-505.
13. Sollid LM, Quiao SW, Anderson RP, et al. Nomenclature and listing of celiac
disease relevant gluten T-cell epitopes restricted by HLA-DQ molecules.
Immunogenetics 2012; 64: 455-460.
14. Enck P, Merlin V, Erckenbrecht JF, et al. Stress effects on gastrointestinal transit
in the rat. Gut 1989; 30: 455-459.
15. Myagmarjalbuu B, Moon MJ, Heo SH, et al. Establishment of a protocol for
determining gastrointestinal transit time in mice using barium and radiopaque
markers. Korean J Radiol 2013; 14: 45-50.
16. Welch MG, Margolis KG, Li Z, et al. Oxytocin regulates gastrointestinal motility,
inflammation, macromolecular permeability, and mucosal maintenance in mice.
Am J Physiol Gastrointest Liver Physiol 2014; 307: G848-G862.
17. De Kauwe A, Chen Z, Anderson RP, et al. Resistance to celiac disease in
humanized HLA-DR3-DQ2-transgenic mice expressing specific anti-gliadin
CD4+ T cells. J Immunol 2009; 182: 7440-7450.
7 Statements on current developments
concerning gluten analysis, clinical and legal
aspects
News from Codex and regulatory affairs
Hertha Deutsch
AOECS Codex Delegate, Austrian Coeliac Society, Vienna
Introduction
AOECS, the Association of European Coeliac Societies, has Observer status in the
Codex Alimentarius Commission since 1992. Information about the organisation, the
duties and all the extensive work of all Codex Committees is published on their
website [1]. In November 2015 and February 2016 two important Codex sessions took
place where issues regarding “gluten-free” were on the agenda: CCNFSDU [2] and
CCMAS [3].
Codex Committee on Nutrition and Foods for Special Dietary Uses
The thirty-seventh Session of the Codex Committee on Nutrition and Foods for
Special Dietary Uses (CCNFSDU) took place from 23 – 27 November 2015 in Bad
Soden, Germany. The Committee was attended by delegates from 66 Member
Countries, one Member Organisation and 36 International Organisations, one of them
was AOECS.
At Agenda item „Matters Referred by the Codex Alimentarius Commission and/or
Other Subsidiary Bodies“, sub-item „Matters for Action“, the CCNFSDU has to
consider the following text of the Codex Document CX/NFSDU 15/37/2: “The 36th Session of the Committee on Methods of Analysis and Sampling
(CCMAS 36)
Examination of “ELISA G12” as a potential additional method for inclusion in
Standard for Foods for Special Dietary Use for Persons Intolerant to Gluten
(CODEX STAN 118-1979)
“19. When considering “ELISA G12” as a potential additional method for inclusion in
CODEX STAN 118-1979, CCMAS 36 noted that any potential endorsement of G12
would be as a Type I procedure and that it would not be possible to have two Type I
methods in the Standard for the same matrices and determination. CCMAS further
observed that if the G12 method for detection of the toxic fraction in gluten harmful
for individuals were added, the provision in the Standard would need to be differentiated to allow for both methods (R5 and G12) to be included as Type I
methods. The Committee noted that G12 had been validated for gluten-free foods, rice
matrices, whereas R5 had been validated for gluten-free foods, maize matrices.
CCMAS 36 recommended that decision in this regard should be taken by CCNFSDU.
20. The Committee is invited to consider the above reply.”
When the item was opened for discussion, two Member countries outside Europe were
in favour of adopting the G12, one Member Organisation was not in favour.
In my comment on behalf of AOECS, I recalled that the threshold for gluten-free was
established based on the results given by the R5. It is not known today whether or not
the G12 gives the same results in all the different kinds of gluten-free foods, e.g.
mixed ingredients from rice and maize or buckwheat or pseudocereals or wheat starch
etc. Further on, there are no data published whether the G12 can detect partly
hydrolysed wheat protein products or beer or malt extracts etc. Regarding oats
products, the G12 shows positive results in some oats samples. Varying results below
or above the threshold of gluten-free in the same food sample will cause severe
difficulties in terms of food labelling and will lead to confusion for coeliacs, food
producers and national food control authorities. Today it is too early to take a decision.
The Chair proposed to establish an ad hoc Working Group with the task to elaborate
questions to CCMAS, which was accepted by the Committee.
The Working Group members considered this item very carefully and elaborated
questions to CCMAS. After some discussion in the Plenary, the Committee approved
the work of the Working Group and the following text for the CCNFSDU report:
“Examination of “ELISA G12” as a potential additional method for inclusion in
Standard for Foods for Special Dietary Use for Persons Intolerant to Gluten
(CODEX STAN 118-1979)
“10. The Committee noted the reply from CCMAS in particular with respect to
validation of the R5 and G12 methods, based on the two matrices, maize and rice but
questioned: which method to adopt for mixed matrices; the comparability of the two
methods (if different results emerge) and the implications for “gluten-free” labelling.
The Committee decided to seek further clarification from CCMAS with the following
request:
Taking into account that the thresholds in CODEX STAN 118-1979 were
established on the basis of the results given by the ELISA R5 Method, can CCMAS
confirm that the results of the two methods (R5 and G12) are fully comparable for
all products covered by the standard, in particular:
o products manufactured from ingredients naturally free of gluten
(e.g. buckwheat, millet, amaranth, quinoa etc.); o products manufactured from gluten-containing ingredients (e.g. partially
hydrolysed wheat protein, wheat starch, malt extract, glucose syrups etc.);
o products based on oats;
o liquid matrices.”
Other important items on the agenda were the Review of the Standard for Follow-up
Formula, the Proposed Draft Definition for Biofortification and a Discussion Paper on
a Standard for Ready-to-use Foods.
Codex Committee on Methods of Analysis and Sampling
The thirty-seventh session of the Codex Committee on Methods of Analysis and
Sampling (CCMAS) took place from 22 – 26 February 2016 in Budapest, Hungary and
was attended by 47 Member Countries, one Member Organisation and Observers from
17 International Organisations, one of them was AOECS.
Before the CCMAS session, the Meeting of International Organisations working in the
field of Methods of Analysis and Sampling (Inter-Agency-Meeting – IAM) took place
on Saturday, 20 February 2016, to consider the items on the agenda of the CCMAS.
Regarding the subject “Comparison of methods for gluten“, the participants published
their statement in the Codex Conference Room Document CRD 4:
„Participants noted that both R5 and G12 based procedures were in use to detect
gluten in food products. Both methods have been validated and adopted by AACCI
with limited scopes based on the range of reference materials and matrices tested. It
was not considered feasible for further validation to be performed without this being
undertaken by the manufacturers of the test kits. Furthermore, the addition of further
matrices and mixed matrices to the scope of application of the tests might be best
carried out by the end-users of the tests where these products occur. IAM members
indicated that an appropriate place to remind users of these matrix restrictions may be
in section 5.2 of the appropriate standard.“
The Codex Working Group on the Endorsement of Methods of Analysis and Sampling
was held on Sunday, 21 February 2016, prior to the Plenary session. Among other
items, the WG discussed also the subject “Examination of “ELISA G12” as a potential additional method for inclusion in the
Standard for Foods for Special Dietary Use for Persons Intolerant to Gluten
(CODEX STAN 118-1979)“ as requested by the Codex Committee on Nutrition and Foods for Special Dietary Uses
(CCNFSDU). After some debate the conclusion was published in the Conference
Room Document CRD 2: “The WG recommended to inform CCNFSDU that (regrettably) the two methods (the
R5 and G12 methods) for the determination of gluten are not comparable. Comparison
data for the two methods are not available, and mixed matrixes are not included in the
scope of either of the methods obtained during their validation. Users confirmed that
the two methods provide different results, and that both methods are used in official
control. The developers of these proprietary methods might be able to provide further
information on the applicability of the methods.”
When the item was discussed in the Plenary session, the recommendation of the
Working Group was supported by the Committee and the following text is in the
CCMAS report:
“The Committee agreed to inform CCNFSDU that the two methods (R5 and G12) for
the determination of gluten are not comparable; that comparability data for the two
methods were not available; and mixed matrices are not included in the scope of either
of the methods obtained during their validation. The developers of these proprietary
methods might be able to provide further information on the applicability of the
methods.”
Since a few years, a further important item is on the agenda of CCMAS:
Development of Procedures/Guidelines for determining equivalency to Type I
Methods
In the past years, an electronic Working Group generally considered the subject of
equivalency to Type 1 Methods and, finally, CCMAS agreed to the following
conclusion as written in the CCMAS report:
“The Committee could not reach consensus on the use and scope of the equivalency
approach and agreed to reconsider this matter in the future when more information
became available. The Committee noted that most of the work in determining
equivalence falls on the Standards Development Organisations (SDOs), and noted the
offer of the SDOs, through the Inter-Agency Meeting (IAM), to look into this matter
and provide recommendations to a future session of CCMAS.”
A further important item on the agenda was
Review and update of Methods in Codex STAN 234-1999
The Committee agreed to continue to work on the review and update of CODEX
STAN 234-1999. All Codex Methods, also the R5 Method, are listed in this Codex
Standard and maybe there will be some debate in the future whether or not to keep the
R5 as Type 1 Method.
References
1. Codex Alimentarius Commission www.codexalimentarius.org/
2. Report of the thirty-seventh session of the Codex Committee on Nutrition and
Foods for Special Dietary Uses, Bad Soden, Germany, 23 - 27 November 2015
3. Report of the thirty-sixth session of the Codex Committee on Methods of Analysis
and Sampling, Budapest, Hungary 22 - 26 February 2016
Considerations concerning methods for gluten
quantitation in foods (R5/G12 ELISA)
Peter Koehler1, Fernando Chirdo2, Thomas Mothes3, Olivier Tranquet4, Katharina A.
Scherf1
1 Deutsche Forschungsanstalt für Lebensmittelchemie, Leibniz Institut, Freising,
Germany
2 Universidad Nacional de La Plata, Facultad de Ciencias Exactas, Instituto de
Estudios Immunologicos y Fisiopatologicos, La Plata, Argentina
3 Institute for Laboratory Medicine, Clinical Chemistry & Molecular Diagnostics,
Medical Faculty of the University and University Hospital, Leipzig, Germany
4 INRA, UR1268 Biopolymers, Interactions, Assemblies, Nantes, France
Introduction
During the sessions and in the coffee breaks there were lively debates on how novel
ELISA methods for the quantitation of gluten could be introduced into practice and
which criteria should be set to acknowledge both established methods and newly
introduced methods. This was particularly related to the R5 and G12 ELISAs and
would also be relevant for test kits introduced into the market in the future. After the
meeting a number of experts met in the lobby of the conference hotel to initiate actions
to deal with this issue. The following statements summarise the suggestions of the
experts and depict a possible way out of this issue.
R5 and G12 ELISAs
The sandwich ELISA based on the R5 monoclonal antibody [1] for the quantitation of
intact gluten was endorsed as a Codex Alimentarius type 1 method [2] and has also
been adopted as AOAC International [3] and AACC International [4] approved
methods. Rice- and maize-based matrices have been used in these studies. A
competitive version of the R5 ELISA has also been approved by both standardisation
organisations [5,6]. In 2008, sandwich and competitive ELISAs based on the G12 (and
A1) monoclonal antibodies were developed [7] and are now commercially available.
The G12 sandwich ELISA has also been validated in an international collaborative
study using rice-based matrices and is approved as AACC International [8] and AOAC
International [9] method.
Codex Alimentarius
Codex analytical methods are being revised every 10 years and revision for the R5
Mendez ELISA is due either in 2017 or 2018. Thus, it is time to discuss how to handle
ELISAs for gluten quantitation regarding approval by the Codex Alimentarius. Based on the matrices used for validation, the R5 method has been recommended for gluten
quantitation in maize matrices and the G12 method for the analysis of rice matrices.
Both methods fulfil the performance requirements for gluten analysis set by the Codex
Standard 118-1979 [10], i.e., a limit of quantitation of 10 mg gluten/kg or less and the
detection of coeliac disease-active epitopes.
At the 37th session CCMAS decided to inform the Codex Committee on Nutrition and
Foods for Special Dietary Uses (CCNFSDU) that the R5 and G12 methods were not
comparable, that comparability data were not available and that the kit producers
should be encouraged to provide more information on the applicability of the methods
[11]. In the following session of CCNFSDU, the committee decided not to include the
ELISA G12 method in the Codex Standard because they noted that the results were not
comparable with the R5 ELISA and recommended to wait for comparability data
provided by the PWG [12].
Concerns
A major concern, in particular of coeliac societies, coeliac disease patients, food
producers and national food control laboratories is the unclear situation if two ELISA
methods for gluten quantitation were endorsed. It can be assumed that each analytical
laboratory would use one ELISA as the default method for gluten quantitation.
Consequently, it would be unclear, if a value obtained by one laboratory with one kit
would be comparable to the value provided by a different laboratory with a different
kit. This would lead to the question how to handle conflicting results from different
laboratories. In general, the possibility of having two type I methods has to be
questioned, because the definition of a type 1 method as “the only method” should
exclude approval of a second type I method. On the other hand, if a proprietary
method fulfilling the performance criteria of the Codex is on the market and has been
approved by suitable collaborative studies, it should not be excluded due to the fact
that another method has already been endorsed by the Codex Alimentarius. Both the
R5 and G12 sandwich ELISAs have been compared in a number of scientific studies.
In summary, the results of these studies strongly suggest that these methods do not
yield comparable results. Typical examples are papers published by Bugyi et al. [13],
Bruins Slot et al. [14] and Scherf [15].
Suggestions
Both ELISAs have gone through several rounds of FAPAS (Food Analysis
Performance Assessment Scheme) proficiency studies. The number of laboratories that
used the R5 ELISA is by far higher compared to the G12 ELISA. However, a
cumulative consideration and statistical evaluation of all available FAPAS data, in
particular those studies in which the same matrices were analysed by both the R5 and
the G12 ELISA are not available to date. Thus, data from FAPAS testing starting at
least from 2012 should be acquired and digitalised. This would enable appropriate statistical analysis and would help to objectively compare the methods. Then
knowledge gaps can be identified and a decision can be made whether additional
analyses of samples (e.g., defined food samples from different categories) are
necessary.
The CCNFSDU noted the reply from CCMAS in particular with respect to validation
of the R5 and G12 methods, based on the two matrices, maize and rice but questioned
which method to adopt for mixed matrices [16]. This could be interpreted by kit
producers that if the matrix would be restrained very clearly and if the method would
be fully validated for this matrix, more than one Codex type I method would be
possible. Consequently, each kit producer would validate its own kit for a specific
matrix and multiple type 1 methods would evolve. This would lead to even more
confusion than with ‘only’ the two current methods based on the R5 and G12
antibodies. This cannot be the goal for the future.
Position of the Prolamin Working Group (PWG)
Therefore, the PWG suggests that ELISA methods should be approved using a
combination of
(1) information on the method that has been used to provide the analytical value,
(2) strict performance criteria and
(3) a pre-defined set of maximum five matrices.
This would be similar to the Standard Method Performance Requirements (SMPR)
published by AOAC International for allergen-containing commodities such as whole
egg, milk, peanut and hazelnut [17].
Performance criteria
Performance criteria include the correct setup and statistical evaluation of validation
studies [18-20] as well as the fulfilment of the requirements for standard method
performance [21]. The minimal performance requirements set in AOAC SMPR
2016.002 [17] for whole egg, milk, peanut and hazelnut can be adapted to gluten, and
the suggested values are given in Table 1. AOAC suggests low recovery rates of 60%,
but the PWG feels that, in general, the recovery range should be between 80 and
120 %, which is in line with Abbott et al. [20]. A recovery rate of 60% should only be
tolerated if repeated analyses with different kits have shown that the low rate is
reproducible among different methods. A range of recovery rates between 50 and
150% for difficult matrices as suggested in [19] should not be accepted with modern
gluten ELISAs. In this context the term “difficult matrix” is not well defined and
should be omitted. Relative standard deviations that have to be tolerated appear to be
rather high (up to 30%), but this seems to be an inherent issue of ELISA methods as
compared to other analytical methods. Nevertheless, method developers should aim to gradually improve these parameters. With respect to limit of detection and limit of
quantitation both R5 and G12 ELISAs perform well [4,5,8].
Table 1. Method performance requirements for gluten ELISAs.

Matrices
Matrixes should not be based on botanical origin (e.g., rice- or maize-based), but on
constituents that most likely affect the interaction of the antibodies with the gluten
antigens. Possible matrixes should be categorised into protein-based, starch-based, fatbased,
polyphenol-rich and fibre-rich foods. Table 2 suggests categories and examples
for foods from each category. Examples are limited to three per category to keep the
number of required analyses in validation studies in a range that can be handled.
Table 2. Suggested matrix categories and examples for foods from each category.

Practical considerations
Kit manufacturers are encouraged to agree on a set of matrixes which should be
comparatively analysed using their methods. In case of conflicting R5/G12 results, in
particular in the concentration range of the 20 mg/kg threshold, the higher concentration
value should be considered relevant in the interest of the coeliac consumers. In
future analyses it should then be avoided having to do two ELISAs. For any analysis
value, the ELISA method that was used should be indicated alongside the results.
Conclusion
The PWG acknowledges that more than one ELISA method for the analysis of gluten
in foods are currently used and that the results of these methods are not comparable.
The group does not support the policy of the Codex Alimentarius to allow approval of more than one type 1 method because this is in contrast to the definition of a type 1
method. The Codex Alimetarius should decide soon how to proceed, if methods are
available that fulfil all performance criteria such as the R5 and G12 ELISAs. The
PWG suggests that performance data of both ELISAs obtained with identical or at
least comparable matrixes should be compared. If existing data is not sufficient,
comparative studies should be carried out on a set of foods suggested in this statement
paper. This could result in a kind of guidebook that suggests specific ELISAs for
specific foods.
References
1. Valdes I, Garcia E, Llorente M, Mendez E. Innovative approach to low level
gluten determination in foods using a novel sandwich enzyme-linked
immunosorbent assay protocol. Eur J Gastroenterol Hepatol 2003; 15: 465-474.
2. Codex Standard 234-1999, 2014. Recommended methods of analysis and
sampling. Codex Alimentarius Commission. Amendment 4.
3. Immer U, Haas-Lauterbach S. Gliadin as a measure of gluten in foods containing
wheat, rye, and barley – enzyme immunoassay method based on a specific
monoclonal antibody to the potentially celiac toxic amino acid prolamin
sequences: collaborative study. J AOAC Int 2012; 95: 1118-1124.
4. Koehler P, Schwalb T, Immer U, et al. AACCI approved methods technical
committee report: collaborative study on the immunochemical determination of
intact gluten using an R5 sandwich ELISA. Cereal Foods World 2013; 58: 36-40.
5. Koehler P, Schwalb T, Immer U, et al. AACCI approved methods technical
committee report: collaborative study on the immunochemical determination of
partially hydrolyzed gluten using an R5 competitive ELISA. Cereal Foods World
2013; 58: 154-158.
6. Lacorn M, Weiss T. Partially hydrolyzed gluten in fermented cereal-based
products by R5 competitive ELISA: collaborative study, first action 2015.05. J
AOAC Int 2015; 98: 1346-1354.
7. Morón B, Cebolla Á, Manyani H, et al. Sensitive detection of cereal fractions that
are toxic to celiac disease patients by using monoclonal antibodies to a main
immunogenic wheat peptide. Am J Clin Nutr 2008; 87: 405-414.
8. Don C, Halbmayr-Jech E, Rogers A, Koehler P. AACCI approved methods
technical committee report: collaborative study on the immunochemical
quantitation of intact gluten in rice flour and rice-based products using G12
sandwich ELISA. Cereal Foods World 2014; 59: 187-193.
9. Halbmayr-Jech E, Rogers A, Don C, Prinster M. Gluten in rice flour and baked
rice products by G12 Sandwich ELISA: First Action 2014.03. J AOAC Int 2015;
98: 103-111.
10. Codex Standard 118-1979, 2015. Codex Standard for Foods for Special Dietary
Use for Persons Intolerant to Gluten. Codex Alimentarius Commission. Revision
1, Amendment 2.
11. Codex Committee of Methods of Analysis and Sampling (CCMAS). REP16/MAS.
Report of the thirty-seventh session, Budapest, Hungary, 22-26 February 2016.
12. Codex Committee on Nutrition and Foods for Special Dietary Uses (CCNFSDU).
REP17/NFSDU. Report of the thirty-eight session, Hamburg, Germany, 5-9
December 2016.
13. Bugyi Z, Torok K, Hajas L, et al. Comparative study of commercially available
gluten ELISA kits using an incurred reference material. Qual Assur Saf Crops
Foods 2013; 5: 79-87.
14. Bruins Slot ID, Bremer MGEG, van der Fels-Klerx I, Hamer RJ. Evaluating the
performance of gluten ELISA test kits: The numbers do not tell the tale. Cereal
Chem 2015; 92: 513-521.
15. Scherf KA. Gluten analysis of wheat starches with seven commercial ELISA test
kits - Up to six different values. Food Anal Methods 2017; 10: 234-246
16. Codex Committee on Nutrition and Foods for Special Dietary Uses (CCNFSDU).
REP16/NFSDU. Report of the thirty-seventh session, Bad Soden, Germany, 23-27
November 2015.
17. AOAC SMPR 2016.002. Standard Method Performance Requirements (SMPRs®)
for Detection and Quantitation of Selected Food Allergens. AOAC International,
March 31, 2016.
18. Appendix D: Guidelines for Collaborative Study Procedures to Validate
Characteristics of a Method of Analysis, Official Methods of Analysis (2016) 20th
Ed., AOAC INTERNATIONAL, Rockville, MD, USA (http://www.eoma.aoac.
org/ app_d.pdf)
19. Abbott M, Godefroy SB, Yeung JM, et al. Validation procedures for quantitative
food allergen ELISA methods: community guidance and best practices. J AOAC
Int 2010; 93: 442-450.
20. Koerner TB, Abbott M, Godefroy SB, et al. Validation procedures for quantitative
gluten ELISA methods: AOAC allergen community and best practices. J AOAC
Int 2013; 96: 1033-1040.
21. Appendix F: Guidelines for Standard Method Performance Requirements, Official
Methods of Analysis (2016) 20th Ed., AOAC INTERNATIONAL, Rockville,
MD, USA (http://www.eoma.aoac.org/app_f.pdf)
8 Perspectives and action plan of the PWG
Peter Koehler
Deutsche Forschungsanstalt für Lebensmittelchemie, Leibniz Institut, Freising, Germany
The Prolamin Working Group executive meeting and joint discussion held on 23
September 2016, led to the decisions and statements outlined below.
Action plan
I. Analytical
Peter Koehler is responsible for the PWG gliadin reference material
(peter.koehler@tum.de).
PWG gliadin will continue to be the reference material supported by the group.
Material for 5 - 10 years is still on stock.
Regarding new reference material, suitable wheat cultivars are currently
identified by the MoniQA initiative. The PWG thinks that flour is not a suitable
reference material and supports a protein sample as reference material. The
flours identified by the MoniQA initiative could be used as starting materials
for isolating protein reference materials for gluten from wheat, rye, and barley.
A statement of the PWG on the use of ELISA kits for the analytical
determination of gluten will be worked out and is published in this book.
Katharina Scherf will be invited to the 2017 meeting to speak on the state-ofthe-
art in gluten quantitation by LC-MS.
II. Clinical
For the symposium of the 2017 meeting the topic “The role of intestinal
microbiota in coeliac disease” has been selected. Three speakers will be invited.
III. Members, Policy
Olivier Tranquet and Rudolf Valenta are new members of the group. René Smulders will replace Luud Gilissen as a group member.
Potential new group members have to be identified in the near future.
Fernando Chirdo is responsible for the website.
This printed, citable book (print run: 250 copies with ISBN number) was made
possible by funding of Dr. SCHÄR GmbH/Srl, (Burgstall, BZ, Italy) and by the
help of Mrs. Anneliese Stoiber and Dr. Gaby Andersen, Deutsche Forschungsanstalt
für Lebensmittelchemie (Freising, Germany). It will be distributed
among leaders of opinion in gluten analysis and clinical medicine. An
electronic version can be downloaded free of charge from the PWG website.
Next meeting: 2016
We are very pleased to announce the venue for our meeting in 2017:
Minden, Germany
Host:
Dr. Markus Brandt
Ernst Böcker GmbH & Co.KG
Ringstraße 55-57
DE-32427 Minden, Germany
Phone: +49 0571 83799-43
Fax: +49 0571 83799-20
E-mail: Markus.Brandt@sauerteig.de
Time: 28 - 30 September 2017
Focus of the meeting:
Role of intestinal microbiota in coeliac disease
Gluten quantitation: LC-MS vs. ELISA
The meeting will be limited to 55 participants and attendance is by
invitation only. Invitations have been sent by March 2017. Registration
deadline will be June 15, 2017.
For registration please contact:
Judith Glöggler
Deutsche Zöliakie-Gesellschaft e.V. (DZG)
Kupferstraße 36
70565 Stuttgart, Germany
Phone: +49 711 45998 49
Fax: +49 711 459981 50
E-mail: judith.gloeggler@dzg-online.de
Very special thanks to the host of this kind invitation!
|
|
List of Participants
GROUP MEMBERS
Prof. Dr. Fernando G. Chirdo
Laboratorio de Investigación en el Sistema Inmune (LISIN)
Facultad de Ciencias Exactas Universidad Nacional de La Plata cc 711
(1900) LA PLATA, ARGENTINA
Phone: +54 221 421 0 497 | 423 0 121 | 423 5 333 (Int 45)
Tele fax +54 221 422 6947
Email: fchirdo@biol.unlp.edu.ar
Prof. Paul J. Ciclitira
King's College London (Division of Diabetes and Nutritional Sciences) The Rayne Institute (KCL). St Thomas' Hospital Westminster Bridge Road. LONDON SE1 7EH, UK/ENGLAND
Phone: +44 207 620 2597 | 207 188 2494 | Telefax +44 207 261 0667
Email: mila.labar_weintrop@kcl.ac.uk (secretary)
Email: paul.ciclitira@kcl.ac.uk
Prof. Dr. Carlo Catassi
(not attending), substituted by
Dr. Tiziana Galeazzi
Università Politecnica delle Marche
Facoltà di Medicina e Chirurgia
Istituto di Clinica Pediatrica
Via Corridoni 11
60123 ANCONA, ITALY
Phone: +39 071 5962834
Fax: +39 071 34270
E-mail: t.galeazzi@univpm.it
Prof. Dr. Peter Köehler
Deut sche Forschungsanstalt für Lebensmittelchemie
Lise-Meitner-Straße : +34 - 85354
FREISING, GER MANY
Phone: +49 81 61 71 29 28 | Tele fax +49 81 61 71 29 70
Email: peter.koehler@tum.de
Prof. Dr. Frits Koning
Leiden Univer sity Medical Center, E3-Q
Department of Immunohaematology and Bloodbank Albinusdreef 2
2333 ZA LEIDEN, THE NETHERLANDS
Phone: +31 71 5266673 | Tele fax +31 71 5265267
Email: fkoning@lumc.nl
Prof. Dr. Knut Lundin
Oslo Universitetssykehus
HF Rikshospitalet
Postboks 495
N-0424 OSLO, NORWAY
Phone: +47 909 80325
Fax: +47 2307 2410Email:
k.e.a.lundin@medisin.uio.no
Prof. Dr. Thomas Mothes
Universitätsklinikum Leip zig A. ö. R.
Institut für Laboratoriumsmedizin, Klinische Chemie und Molekulare Diagnostik - Liebigstraße : 27 - 04103
LEIPZIG, GERMANY
Phone: +49 341 97 22251 | Tele fax +49 341 97 22329
Email: mothes@medizin.uni-leipzig.de
Dr. René Smulders
Plant Research International (PRI)
Wageningen University
Droevendaalsesteeg 1
6708 PB WAGENINGEN,
THE NETHERLANDS
Phone: +31 317 480840
Email: rene.smulders@wur.nl
Dr. Olivier Tranquet
INRA
Rue de la Géraudière BP 71627
44316 NANTES CEDEX 3, FRANCE
Phone: +33 2 40675027
Fax: +33 240675025
E-mail: olivier.tranquet@nantes.inra.fr
Prof. Dr. Riccardo Troncone
Department of Pediatrics and European
Laboratory for the Investigation of
Food-Induced Diseases
University of Naples “Federico II”
Via Pansini, 5
80131 NAPLES, ITALY
Phone: +39 081 7463383
Fax: +39 081 5469811
E-mail: troncone@unina.it
Prof. Dr. Detlef Schuppan
(not attending), substituted by
Dr. Victor Zevallos
I. Medizinische Klinik und Poliklinik
Universitätsmedizin der Johannes
Gutenberg-Universität Mainz
Institut für Translationale Medizin
Obere Zahlbacher Str. 63
55131 MAINZ, GERMANY
Phone: +49 6131 17 9783
Fax: +49 6131 17 9988
Email: zevallos@uni-mainz.de
HOSTS
Prof. Dr. Cristina Molina Rosell
Institute of Agrochemistry and Food
Technology (IATA-CSIC)
Avenida Agustin Escardino 7
46980 PATERNA, VALENCIA,
SPAIN
Phone: +34 963 900022
Fax : +34 963 636301
E-mail: crosell@iata.csic.es
Ms. Maria Vicenta San Eustaquio
Tarazona
Institute of Agrochemistry and Food
Technology (IATA-CSIC)
Avenida Agustin Escardino 7
46980 PATERNA, VALENCIA,
SPAIN
Phone: +34 963 900022
Fax : +34 963 636301
E-mail: marivise@iata.csic.es
INVITED SPEAKER
Dr. Andreas Frey
Forschungszentrum Borstel
Leibniz-Zentrum für Medizin und
Biowissenschaften
Parkallee 1-40
23845 BORSTEL, GERMANY
Phone: +49 4537 188 - 5620
Fax: +49 (0) 4537 88-6930
E-mail: afrey@fz-borstel.de
Dr. Eva Helmerhorst
Boston University
Department of Molecular and Cell
Biology
700 Albany Street
BOSTON, MA 02118
USA
E-mail: helmer@bu.edu
Prof. Dr. Yolanda Sanz
Institute of Agrochemistry and Food
Technology (IATA-CSIC)
Avenida Agustin Escardino 7
46980 PATERNA, VALENCIA,
SPAIN
E-mail: yolsanz@iata.csic.es
GUESTS
Mrs. Tova Almlöf
Semper AB
Semper AB Box 1101
SE 17222 SUNDBYBERG, SWEDEN
Phone : +46 767232862
E-mail: tova.almlof@semper.se
Mrs. Sofia Beisel
Deutsche Zöliakiegesellschaft e.V.
Kupferstr 36
70565 STUTTGART, GERMANY
Phone: +49 711 45998115
Fax: 49 711 459981 50
E-mail: sofia.beisel@dzg-online.de
Dr. Markus Brandt
Ernst Böcker GmbH & Co KG
Ringstrasse 55-57
32427 MINDEN, GERMANY
Phone: +49 571 837990
Fax: +49 571 8379920
E-mail: markus.brandt@sauerteig.de
Dr. Maria Angeles Bustamente
Universidad del Pais Vasco
Euskal Herrika Unibertsitatea
Universität des Baskenlandes / EHU
Barrio Sarriena s/n Neighborhood
Sarriena s / n
48940 LEIOA; BIZKAIA, SPAIN
E-mail: marian.bustamente@ehu.eus
Dr. Angel Cebolla-Ramirez
Biomedal, SL
Avenida Américo Vespucio, 04.05
Geschoss 1, Modul 12,
41092 SEVILLA, SPAIN
Phone: +34 954 08 12 76
Fax: +34 954 08 12 79
E-mail: acebolla@biomedal.com
Mr. Henrik Dahlquist
Fria Gluten Free
Fältspatsgatan 12
43021 VÄSTRA FRÖLUNDA,
SWEDEN
Phone: +46 70 654 600
E-mail: henrik.dahlquist@fria.se
Dr. Johan De Meester
Cargill R&D Centre Europe
Havenstraat 84
B-1800 VILVOORDE, BELGIUM
Phone: +32 473997653
E-mail: Johan_De_Meester@cargill.com
Mrs. Hertha Deutsch
Österreichische Arbeitsgemeinschaft
Zöliakie
Anton Baumgartner Straße 44/C5/2302
1230 VIENNA, AUSTRIA
Phone: +43 166 71887
E-mail: hertha.deutsch@chello.at
Mrs. Angela Durá de Miguel
Institute of Agrochemistry and Food
Technology (IATA-CSIC)
Avenida Agustin Escardino 7
46980 PATERNA, VALENCIA,
SPAIN
E-mail: andudemi@iata.csic.es
Mr. Richard Fielder
Bio-Check (UK)
Spectrum House, Llys Edmund Prys,
St. Asaph Business Park
LL17 0JA ST. ASAPH, UK
Phone: +44 1745 335165
Fax: +44 1745 582867
E-mail: richard@biocheck.uk.com
Mrs. Raquel Garzón Lloria
Institute of Agrochemistry and Food
Technology (IATA-CSIC)
Avenida Agustin Escardino 7
46980 PATERNA, VALENCIA, SPAIN
E-mail: r.garzon@iata.csic.es
Mrs. Anna Gibert Casamada
Associació Celíacs de Catalunya
Independencia, 257
08026 BARCELONA, SPAIN
E-mail: annagibertc@gmail.com
Ms. Eugenia Yaiza Benavent Gil
Institute of Agrochemistry and Food
Technology (IATA-CSIC)
Avenida Agustin Escardino 7
46980 PATERNA, VALENCIA, SPAIN
E-mail: yaizabenavent@gmail.com
Dr. Gyöngyvér Gell
MTA Centre of Agricultural Research
Department of Applied Genomics
Brunszvik 2
2462 MARTONVÁSÁR, HUNGARY
Phone: +36 225 69521
Fax: +36 225 69514
Email: gell.gyongyver@agrar.mta.hu
Mr. Phil Goodwin
Bio-Check (UK)
Spectrum House, Llys Edmund Prys,
St. Asaph Business Park
LL17 0JA ST. ASAPH, UK
Phone: +44 1745 335165
Fax: +44 1745 582867
E-mail: phil@biocheck.uk.com
Dr. Thomas Grace
Bia Diagnostics
480 Hercules Dr.
5446 COLCHESTER, VT, USA
Phone: +1 802 540 0148
E-mail:
thomasgrace@biadiagnostics.com
Mr. Daniele Grano
Dr. Schär AG /SPA
Winkelau 9
39014 BURGSTALL, ITALY
E-mail: Daniele.Grano@drschaer.com
Mrs. Mia Hallgren
Swedish Food Agency
P.O. Box 622
SE-751 26 UPPSALA, SWEDEN
E-mail: mia.hallgren@slv.se
Mrs. Katri Hautanen
Fria Gluten Free
Fältspatsgatan 12
421 30 VÄSTRA FRÖLUNDA, SWEDEN
Phone: +46 70 5156683
E-mail: katri.hautanen@fria.se
Mr. Xin Huang
University of Helsinki
Department of Food and
Environmental Sciences
Agnes Sjöbergin katu 2, PL66
14 HELSINKI, FINLAND
Phone: +358 451210203
E-mail: xin.huang@helsinki.fi
Mrs. Jasmin Kraus
Romer Labs Division Holding GmbH
Erber Campus 1
3131 GETZERSDORF, AUSTRIA
Phone: +43 2782 803 0
E-mail: jasmin.kraus@romerlabs.com
Dr. Götz Kröner
Hermann Kröner GmbH
Lengericher Str. 158
49479 IBBENBÜREN, GERMANY
Phone: +49 5451 9447 11
Fax: +49 5451 9447 811
E-mail: kroener@kroener-staerke.de
Mrs. Barbara Lexhaller
Deutsche Forschungsanstalt
für Lebensmittelchemie
Lise-Meitner-Str. 34
85354 FREISING, GERMANY
Phone: +49 8161 71 2926
E-mail: Barbara.Lexhaller@Lrz.tum.de
Mrs. Stelle Lindeke
R-Biopharm AG
An der neuen Bergstraße 17
64297 DARMSTADT, GERMANY
Phone: +49 6151 8102 92
E-mail: s.lindeke@r-biopharm.de
Dr. Maria Carmen Mena Valverde
National Center of Biotechnology, CSIC
28049 MADRID, SPAIN
Phone: +34 915854670
Fax: +34 915854506
Email: mcmena@cnb.csic.es
Dr. Luisa Novelino
Fondazione Celiachia
Via Caffaro, 10
16124 GENOVA, ITALY
Phone: +39 010 8449406
Email: lnovellino@celiachia.it
Mrs. Ombretta Polenghi
Dr. Schär R&D Centre
c/o AREA Science Park
Padriciano, 99
I-34149 TRIESTE, ITALY
Phone: +39 040 3755 382
Email: ombretta.polenghi@drschaer.com
Dr. Elena Quesada Hernández
Biomedal, SL
Avenida Américo Vespucio, 04.05
Geschoss 1, Modul 12,
41092 SEVILLA, SPAIN
E-mail: elena.quesada@biomedal.com
Mrs. Catherine Remillieux-Rast
Association Française des Intolérants
au Gluten (AFDIAG)
23 Rue de Venise
78740 VAUX-SUR-SEINE, FRANCE
Phone: +33 681270911
Fax: +33 130993668
E-mail: c.remillieux_rast@yahoo.fr
Dr. Adrian Rogers
Romer Labs UK Ltd.
The Health Business and
Technical Park
WA74QX RUNCORN, CHESHIRE, UK
Phone: +44 845519 0510
Email: adrian.rogers@romerlabs.com
Dr. Cristina Romero
INGENASA
C/Hermanos García Noblejas, 39
28037 MADRID, SPAIN
Phone: +34 91 368 05 01
Fax: +34 91 408 75 98
E-mail: cromero@ingenasa.com
Mr. Nermin Sajic
EuroProxima B.V.
Beijerinckweg 18
6827 BN ARNHEM, THE
NETHERLANDS
Phone: +31 263630364
E-mail: info@europroxima.com
Dr. Martin Salden
EuroProxima B.V.
Beijerinckweg 18
6827 BN ARNHEM, THE
NETHERLANDS
Phone: +31 26 3630364
Email: m.salden@noviosmart.eu
Prof. Hannu Salovaara
University of Helsinki
Department of Food and
Environmental Sciences
Agnes Sjöbergin katu 2, PL66
14 HELSINKI, FINLAND
E-mail: hannu.salovaara@helsinki.fi
Dr. Vikas Kumar Sarna
Dep. of Immunology Hospital-
Rikshospitalet
NORWAY
E-mail: vikas.sarna@medisin.uio.no
Dr. Katharina Scherf
Deutsche Forschungsanstalt für
Lebensmittelchemie
Lise Meitner-Strasse 34
85354 FREISING, GERMANY
Phone: +49 8161712927
Fax: +49 8161712970
E-mail: katharina.scherf@lrz.tum.de
Dr. Juan Ignacio Serrano-Vela
Asociacion de Celiacos de Madrid
Calle Lanuza 19-bajo
28028 MADRID, SPAIN
Phone: +34 917130147
Fax: +34 917258059
E-mail: nachoserrano@celiacosmadrid.org
Dr. Edurne Simón
University of the Basque Country
UPV/EHU
Paseo de la Universidad, 7
1006 VITORIA-GASTEIZ, SPAIN
Phone: +34 945013069
Fax: +34 945013014
E-mail: edurne.simon@ehu.es
Mrs. Gry Skodje
Oslo University Hospital
OSLO, NORWAY
Email: g.i.skodje@gmail.com
Dr. Tuula Sontag-Strohm
University of Helsinki
Agnes Sjöbergin katu 1
00014, HELSINKI, FINLAND
Phone: +358 504487467
E-mail: tuula.sontag-strohm@helsinki.fi
Mrs. Pauline Titchener
Neogen Europe Ltd.
The Dairy School, Auchincruive
KA6 5HU AYR, SCOTLAND, UK
Phone: +44 1292 525 600
Fax: +44 1292 525 601
E-mail: p.titchener@neogeneurope.com
Dr. Angel Venteo
INGENASA
C/Hermanos García Noblejas nº 41
28037 MADRID, SPAIN
Phone: +34 91 3680501
Fax: +34 91 4087598
E-mail: aventeo@ingenasa.com
Dr. Paul Wehling
Medallion Labs
9000 Plymouth Avenue North,
MINNEAPOLIS, MN 55427, U.S.A.
Phone: +1 763 764 4360
E-mail: paul.wehling@genmills.com
Dr. Thomas Weiss
R-Biopharm AG
An der neuen Bergstrasse 17
64297 DARMSTADT, GERMANY
Phone: +49 6151 8102 186
E-mail: t.weiss@r-biopharm.de
Mrs. Maren Wiese
Hermann Kröner GmbH
Lengericher Straße 158
49479 IBBENBÜREN, GERMANY
Phone: +49 5451 9447 12
Fax: +49 5451 9447 812
E-mail: wiese@kroener-staerke.de
Impressum
Proceedings of the 30th Meeting
WORKING GROUP
on PROLAMIN ANALYSIS and TOXICITY
22 - 24 September 2016
Valencia, Spain
This work including all parts is subject to copyright. All rights are reserved and any
utilisation is only permitted under the provisions of the German Copyright Law.
Permissions for use must always be obtained from the publisher. This is in particular
valid for reproduction, translation, conversion to microfilm and for storage or
processing in electronic systems.
Scientific Organisation
Prof. Dr. Peter Koehler
Deutsche Forschungsanstalt für Lebensmittelchemie
Lise-Meitner-Str. 34, 85354 FREISING, GERMANY
Phone: +49 8161 712928; Fax: +49 8161 712970
E-mail: peter.koehler@tum.de
Host
Prof. Dr. Cristina M. Rosell
Institute of Agrochemistry and Food Technology (IATA-CSIC)
Avenida Agustin Escardino 7, 46980 PATERNA, VALENCIA, SPAIN
Phone: +34 963 900022, Fax: +34 963 636301
E-mail: crosell@iata.csic.es
Cover picture* and picture of participants
Thomas Mothes
© Peter Koehler
ISBN: 978-3-00-055981-5
|


